Natural Killer Cells
Learning Objectives
• Identify the phenotypic characteristics of natural killer (NK) cells
• Recognize the mechanism by which NK cells recognize aberrant or infected cells
• Differentiate between the two major classes of activation and inhibition molecules of NK cells
• Identify the major ligand CD94–NKG2 complex
• Compare and contrast the physiologic roles of the subpopulations of NK cells
• Contrast the roles of NK cells in viral infections and tumor surveillance
• Explain the mechanism involved in antibody-dependent cellular cytotoxicity (ADCC)
• Compare and contrast the physiologic roles of NK cells and NKT cells
• Recognize the phenotypic characteristics of NKT cells
• Recognize the two major subsets of NKT cells
• Discuss the role of alpha galactosyl ceramide (α-GalCer) in NKT cell stimulation
• Compare the roles of IL-12 and IL-13 in tumor immunity
• Explain the relationship between type I NKT cells and asthma
• Identify the roles of type II NKT cells in viral and autoimmune diseases
• Identify the nature of the genetic defect in Chediak-Higashi syndrome (CHS)
Introduction
Natural killer (NK) cells are defined by the expression of CD16 and CD56 surface molecules. In peripheral blood, two populations of NK cells are present. One population comprises large granular lymphocytes that are phenotyped as CD3–, CD16+, CD56+, and CD94+ cells. Since they do not express T or B cell markers, they are considered to be of a third lymphocyte lineage. The second population of NK cells comprises T cells that express both T cell and NK cell markers (CD3+, CD16+, CD56+ or CD3+, CD16–, CD56+). These cells are known as natural killer T cells (NKT cells) to differentiate them from large granular lymphocytes.
NK and NKT cells have different functions. NK cells are part of the innate cell-mediated response to infected or tumor cells. They recognize and lyse cells with downregulated human leukocyte antigen (HLA) class I molecules. In contrast to classic NK cells, NKT cells have limited cytolytic capability and require antigen presentation. Emerging evidence also suggests that NKT cells are involved in immediate allergic reactions, suppression of auto-reactive lymphocytes, and tumor immunity. Activation or inhibition of NK and NKT cells is dictated by interactions between cell receptors and HLA class IB molecules or CD1 molecules on target cells.
Natural Killer Cell Receptors
CD94–NKG2 Receptors
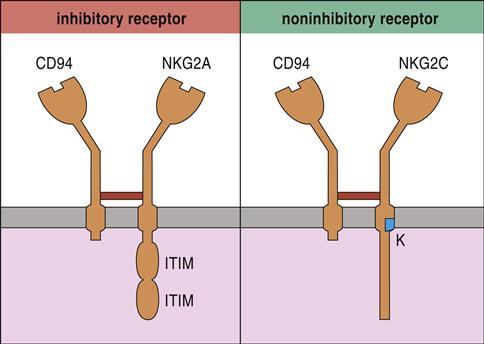
The most studied NK cell receptor is CD94—a type 2 “lectin-like” receptor expressed on both NK cells and cytotoxic T cells (CTLs). To create an active receptor, CD94 pairs with NKG2A, B, C, E, or F. NK cell function is dictated by different pairings between CD94 and NKG family members. For example, NKG2A and NKG2B dimerize with CD94 and inhibit NK cell function. Conversely, CD94–NKG2C dimers activate NK cells (Figure 21-1).
Killer Cell Immunoglobulin-Like Receptors
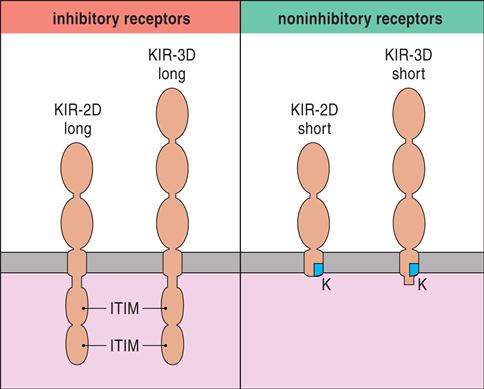
Killer cell immunoglobulin-like receptors (KIRs) are encoded by a family of 15 polymorphic genes and two pseudogenes on chromosome 19. KIRs are membrane anchor proteins that possess either two (KIR2DS) or three (KIR3DS) immunoglobulin domains (Figure 21-2).
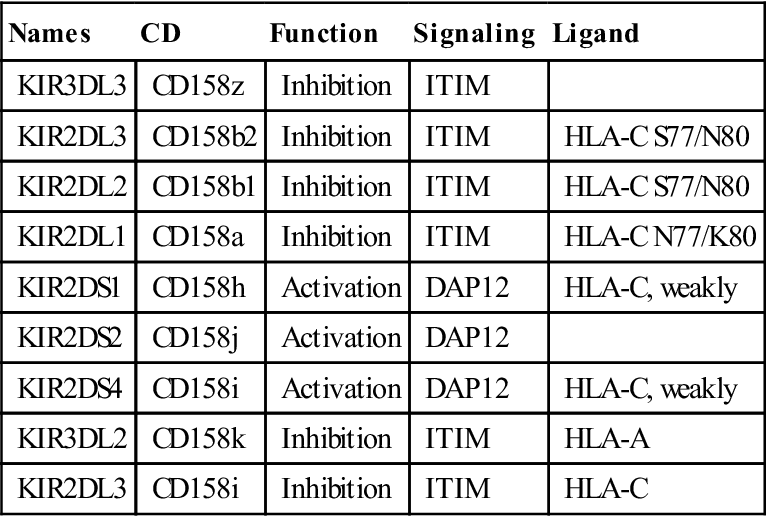
Within each subfamily, KIRs that inhibit or activate NK cells are present (Table 21-1).
Table 21-1
Immune Receptors Encoded by Genes in the Leukocyte Receptor Complex Region
< ?comst?>
| Names | CD | Function | Signaling | Ligand |
| KIR3DL3 | CD158z | Inhibition | ITIM | |
| KIR2DL3 | CD158b2 | Inhibition | ITIM | HLA-C S77/N80 |
| KIR2DL2 | CD158b1 | Inhibition | ITIM | HLA-C S77/N80 |
| KIR2DL1 | CD158a | Inhibition | ITIM | HLA-C N77/K80 |
| KIR2DS1 | CD158h | Activation | DAP12 | HLA-C, weakly |
| KIR2DS2 | CD158j | Activation | DAP12 | |
| KIR2DS4 | CD158i | Activation | DAP12 | HLA-C, weakly |
| KIR3DL2 | CD158k | Inhibition | ITIM | HLA-A |
| KIR2DL3 | CD158i | Inhibition | ITIM | HLA-C |
< ?comen?>< ?comst1?>
< ?comst1?>
< ?comen1?>
Modified from Cooper MD, Lewis LL, Conley, ME, et al: Immunodeficiency disorders. American Society of Hematology Education Program, pp. 314–330, 2003. Hematology 2003, The American Society of Hematology.
Target Cell Recognition Molecules
NK cells do not detect aberrant or infected cells. Rather, they recognize cells that lack HLA class I molecules. This observation has led to the development of the “missing self” hypothesis of NK cell activation. The hypothesis suggests that NK cells can kill normal cells but are prevented from doing so by the presence of inhibitory factors such as HLA class I markers. Downregulation of class I molecules allows NK cells to engage other molecules that either activate or inhibit the NK cell.
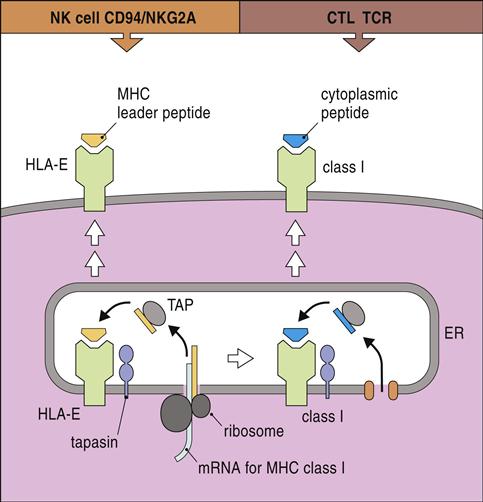
HLA-E, a nonclassic HLA locus, is the natural ligand for CD94–NKG2A and B complexes. HLA-E binds the peptide leader sequence from HLA-G. The HLA-G sequence is encoded upstream of the open reading frame for the α-chain. In the cytoplasm, the N-terminal fragment of the leader sequence is released after cleavage by signal peptidases and endoplasmic reticulum (ER) proteases. The small leader peptide is loaded into the binding groove in the ER and transported to the cell surface (Figure 21-3).
KIRs also interact with HLA-C alleles to activate or inhibit NK cell function. Alleles that inhibit NK cell function have dimorphic polymorphisms at positions 77 and 80 of the binding cleft. The HLA-C1 group variants with serine at position 77 and asparagines at position 80 are HLA-Cw∗102, ∗304, ∗0702, and Cw∗0801. HLA-C alleles with these polymorphisms bind to the NK inhibitory receptors KIR2DL2 and KIR2DL3 and occasionally to the activating factor KIR2DS2. A second variant or HLA-C2 type 2 has asparagine at position 77 and lysine at position 80 (HLA-Cw∗0201, ∗0401, ∗0501, ∗0601, and ∗1503) and binds KIR2DL1, which inhibits the NK response.
Intracellular Signaling by Natural Killer Cells
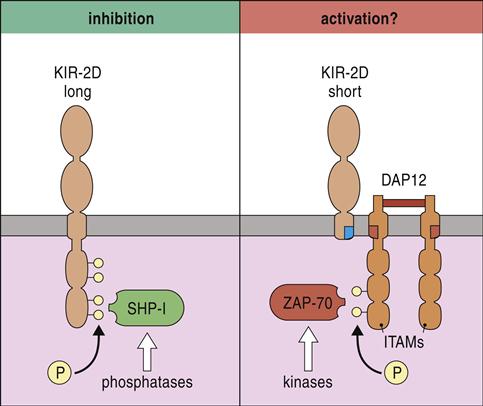
Inhibition or activation of NK cells depends on the presence or absence of long cytoplasmic tails containing immunoreceptor tyrosine-based inhibitory motifs (ITIMs). Receptors with ITIMs are usually phosphorylated and recruit phosphatases such as SHIP-1 (SH2 [Src homology 2]-containing inositol phosphatase-1) and SHIP-2 that inhibit intracellular signaling. In contrast, receptors that lack long cytoplasmic tails associate with DNAX-activating protein of 10Kda (DAP10) and DAP12 proteins, which act as immunoreceptor tyrosine-based activation motifs (ITAM). Phosphorylation of zeta-chain-associated protein kinase (ZAP) initiates a signaling cascade, which activates NK cells (Figure 21-4).
Subpopulations of Natural Killer Cells and Target Cell Lysis
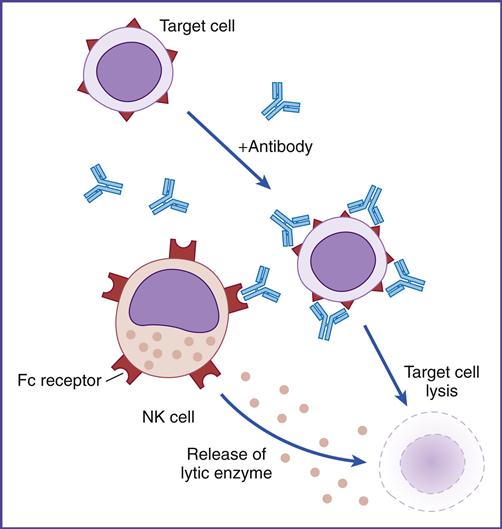
Using flow cytometry, NK cell subpopulations can be indentified on the basis of the numbers of CD56 glycoproteins on the cell surface. Approximately 95% of NK cells express low numbers of CD56 and are termed CD56dim. These cells lyse target cells by using perforin and granzymes. CD56dim cells also express CD16, which is a low-affinity Fc gamma (Fc-γ) receptor. Engagement of the NK cell Fc receptor by antibodies bound to target cells initiates the release of perforin and granzymes. This mechanism is known as antibody-dependent cellular cytotoxicity (ADCC) or killer (K) cell cytotoxicity (Figure 21-5).
Five percent of NK cells express large numbers of CD56 (CD56bright). This population does not contain perforin granules and is not cytotoxic. The CD56bright cells secrete interferon gamma (IFN-γ) and other proinflammatory cytokines such as IL-5, IL-13, and TNF-α.
Function of Natural Killer Cells
NK cells play a critical role in the innate response to viruses, tumors, and fungal infections. During the early stages of a viral infection, NK cells are the primary effector cells. In some instances, NK cells may supplant the T cell response during infections by viruses that downregulate the HLA class I marker expression.
NK cells also play a critical role in the defense against tumors. Because of their presence in peripheral blood, NK cells intercept and kill cancer cells before they can metastasize to distant sites in the body.
NK cells play an indirect role in the termination of opportunistic Candida albicans and Aspergillus infections in immunosuppressed individuals. Production of INF-γ by noncytotoxic NK cells activates macrophage and effector lymphocytes and recruits neutrophils to kill the fungi.
Lymphokine-Activated Killer Cells
When NK cells are stimulated by IL-2 or INF-γ, a dramatic increase occurs in the synthesis of perforin and granzymes. These cells, known as lymphokine-activated killer (LAK) cells, can lyse a broad range of tumor types, including those which are normally resistant to NK cell lysis.
LAK cells have been used in tumor immunotherapy with varying degrees of success. Infusion of autologous LAK cells and IL-2 into patients with some end-stage metastatic cancers was shown to reduce or eradicate the tumor. Currently, LAK cells are being evaluated for the treatment of neuroblastoma and other recurrent brain tumors.
Natural Killer T Cells
On the basis of the structure of the T cell receptor, NKT cells are subdivided into type I and type II cells. Type I NKT (CD3+, CD16+, CD56+) cells have an invariant T cell receptor alpha (TCR-α) chain rearrangement (Valpha24:Jalpha18 chains). These cells recognize a self-glycolipid antigen called alpha galactosylceramide (α-GalCer) presented by CD1 molecules. Antigen-activated type I NKT cells release proinflammatory cytokines such as INF-γ, IL-2, IL-4, IL-12, and IL-13, which can modulate Th1 and Th2 response.
Type II NKT (CD3+, CD16–, CD56+) cells lack the TCR invariant TCR (Valpha24–) α-chain rearrangement but still recognize antigens presented by CD1. The self-antigens, presented by CD1, however, differ from those recognized by type I NKT cells. The physiologic role of type II NKT cells has not been well studied. In animal models, type II NKT cells can augment or inhibit an immune response.
Natural Killer T Cells in Disease
In the immune response to tumors, type I NKT cells can be protective or suppress the immune response. α-GalCer–stimulated type I NKT cells are indirectly involved in the defense against tumors. Type I NKT cells produce INF-γ, which increases CD8 and NK cell lysis of target cells. Cytokines from type I NKT cells also stimulate dendritic cells to produce IL-12, which is important in tumor immunity. IL-12 confers migratory properties to T cells and prepares tumor masses to accept the migrating T cells. In contrast, the production of IL-13 from type I NKT cells has been implicated in the suppression of the anti-tumor response.
Type I NKT cells may also play a critical role in immediate allergic reactions. Patients with severe asthma or uncontrolled asthma have elevated numbers of type I NKT cells in bronchial and alveolar fluids. These cells produce high levels of IL-4, IL-13, and INF-γ. IL-4 production by type I NKT cells may direct the Th2 response and the production of immunoglobulin E (IgE). Although the mechanism is unknown, IL-13 plays a critical role in the development of airway hyperresponsiveness, airway remodeling, mucus secretion, and recruitment of eosinophils.
Type II NKT cells are involved in the pathophysiology of some viral infections and autoimmune diseases. In hepatitis C infections, type NKT II cells produce cytokines that stimulate a Th1 response required for resolution of the infection. In ulcerative colitis, type II NKT cells produce IL-13, which shifts the response to Th2 cells and antibody production. IL-13 acts directly on B cells, stimulating proliferation and isotypic switching to IgG. Autoantibodies directed at neutrophil granules, the portal tract, and colon cells are common in patients with inflammatory bowel disease.
Immunodeficiencies
Chediak-Higashi Syndrome
Chediak-Higashi syndrome (CHS) is a rare autosomal disorder of childhood. Children usually present with hypopigmentation of the eyes, silver hair, platelet dysfunction with prolonged bleeding times, and peripheral neuropathy.
Individuals with CHS have a defective gene called LYST or CHS1, which is responsible for the synthesis, maintenance, and storage of intracellular granules containing perforin and granzymes. NK cells from these patients can bind to target cells but cannot kill cells using the perforin–granzyme pathway.
The inability to form granules is not restricted to NK cells. Neutrophils (azurophilic granules), leukocytes (lysosomes), and melanocytes (melanosomes) also have defective granules. Most patients have a high risk for Epstein-Barr virus (EBV) infections and exhibit a life-threatening, nonmalignant lymphoma-like infiltration of multiple organs. Most patients die from repeated bacterial infections by Staphylococcus aureus, Streptococcus pyogenes, or Pneumococcus species.
Treatment for Chediak-Higashi Syndrome
Bone marrow transplantation (BMT) from an HLA-matched sibling is the therapy of choice. Without a BMT, most individuals usually die before 10 years of age. Before transplantation, patients are maintained on a therapy regimen that controls infections and prevents the spread of viral infections (Table 21-2). Although BMT resolves the immunologic issue, it has no effect on the neurologic and pigmentation problems.
Table 21-2
Treatment for Chediak-Higashi Syndrome
| Drug | Action |
| Acyclovir | Causes deoxyribonucleic acid (DNA) chain termination during viral replication |
| Pooled immunoglobulin | Provides passive immunity to common bacteria and viruses. Neutralizes antimyelin antibodies involved in peripheral neuropathy |
| Interferon alpha-2a and -2b | Prevents spread of viral infections, inhibits proliferation of lymphocytes, and downregulates immune responses |
| Vincristine | Decreases reticuloendothelial function |
| Vinblastine | Disrupts the formation of the mitotic spindle and arrests cell proliferation at metaphase |
| Colchicine | Decreases leukocyte motility and phagocytosis in inflammatory responses |
Summary
• Natural killer (NK) cells are large granular lymphocytes that are neither T cells nor B cells.
• Small populations of T cells also act as NK cells and are designated as NKT cells.
• NKT cells recognize self-antigens presented by CD1 molecules.
• NKT cells are involved in the pathophysiology of asthma, hepatitis C, and ulcerative colitis.