Molecular Basis for Embryonic Development
One of the most important realizations has been the conservatism of the genes that guide development. Sequencing studies have shown remarkably few changes in the nucleotide bases of many developmentally regulated genes that are represented in species ranging from worms to Drosophila to humans. Because of this phylogenetic conservatism, it has been possible to identify mammalian counterparts of genes that are known from genetic studies to have important developmental functions in other species (Box 4.1).* It is also clear that the same gene may function at different periods of development and in different organs. Such reuse greatly reduces the total number of molecules that are needed to control development. Before and after birth, specific genes may be expressed in normal and abnormal processes. One of the principal themes in contemporary cancer research is the role of mutant forms of developmentally important genes (e.g., proto-oncogenes) in converting normal cells to tumor cells.
Fundamental Molecular Processes in Development
From a functional standpoint, many of the important molecules that guide embryonic development can be grouped into relatively few categories. Some of them remain in the cells that produced them and act as transcription factors (Fig. 4.2). Transcription factors are proteins possessing domains that bind to the DNA of promoter or enhancer regions of specific genes. They also possess a domain that interacts with RNA polymerase II or other transcription factors and consequently regulates the amount of messenger RNA (mRNA) produced by the gene.
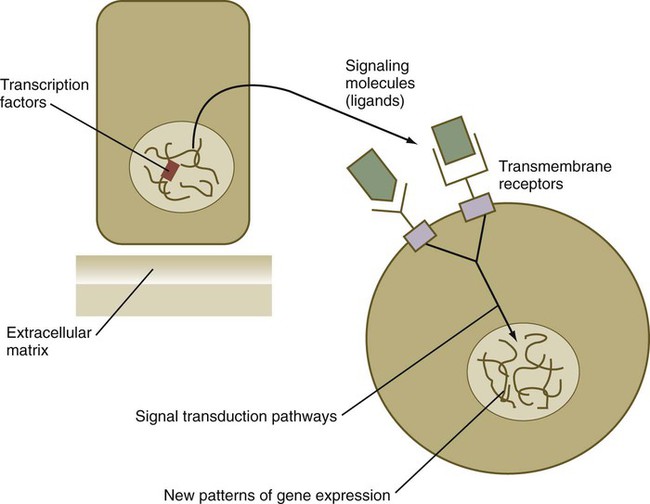
Transcription Factors
Homeobox-Containing Genes and Homeodomain Proteins
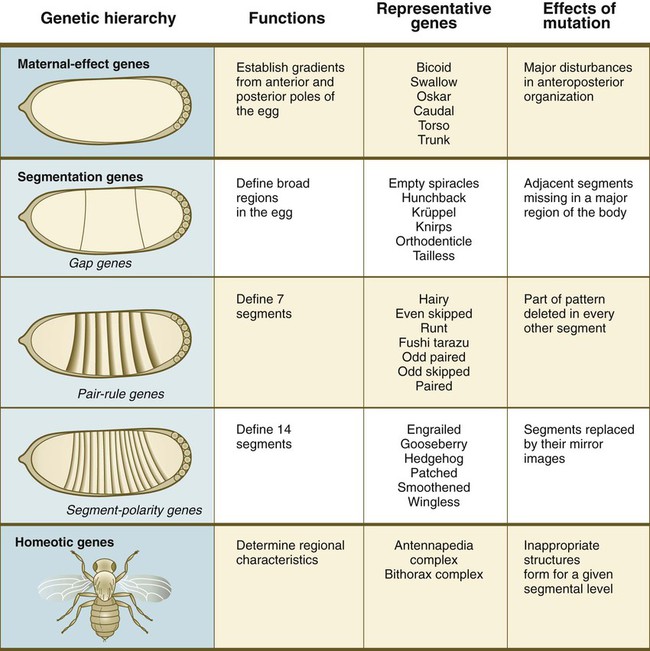
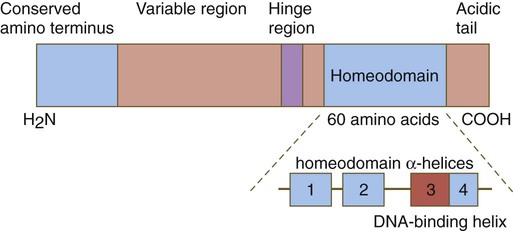
One of the most important types of transcription factors is represented by the homeodomain proteins. These proteins contain a highly conserved homeodomain of 60 amino acids; a homeodomain is a type of helix-loop-helix region (Fig. 4.3). The 180 nucleotides in the gene that encode the homeodomain are collectively called a homeobox. Homeobox regions were first discovered in the homeotic genes of the antennapedia and bithorax complex in Drosophila (see Fig. 4.1), hence their name. This designation sometimes confuses students because, since their initial description, homeoboxes have been found in several more distantly related genes outside the homeotic gene cluster. Many other gene families contain not only a homeobox but also other conserved sequences (Fig. 4.4).
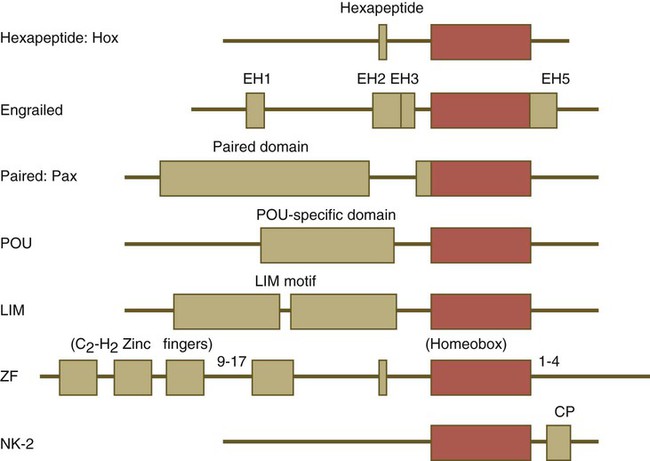
Names of the different classes of genes are listed on the left. The red boxes represent the homeobox within each gene class. The other boxes represent conserved motifs specific to each class of genes. (Modified from Duboule D, ed: Guidebook to the homeobox genes, Oxford, 1994, Oxford University Press.)
HOX Genes
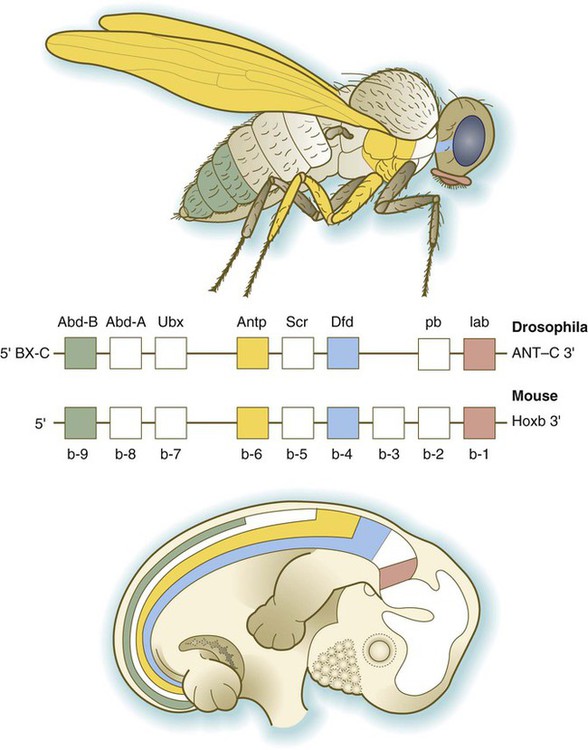
The Drosophila antennapedia-bithorax complex consists of 8 homeobox-containing genes located in 2 clusters on one chromosome. Mice and humans possess at least 39 homologous homeobox genes (called Hox genes in vertebrates [HOX in humans]), which are found in 4 clusters on 4 different chromosomes (Fig. 4.5). The Hox genes on the 4 mammalian chromosomes are arranged in 13 paralogous groups.
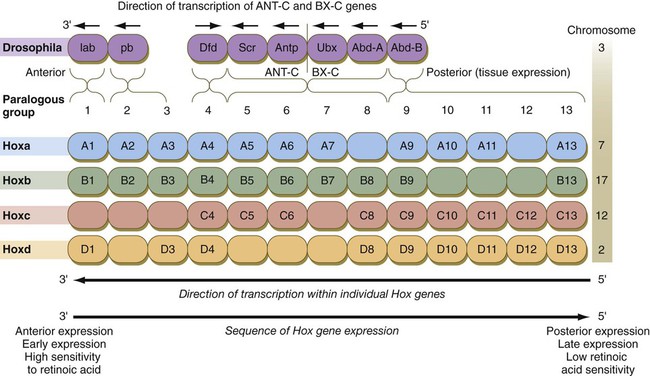
Genes on the 3′ ends of each of the complexes are expressed earlier and more anteriorly than those on the 5′ end (right). (Based on Scott MP: Cell 71:551-553, 1992.)
Vertebrate Hox genes play a prominent role in the craniocaudal segmentation of the body, and their spatiotemporal expression proceeds according to some remarkably regular rules. The genes are activated and expressed according to a strict sequence in the 3′ to 5′ direction, corresponding to their positions on the chromosomes. Consequently, in Drosophila and mammals, 3′ genes are expressed earlier and more anteriorly than are 5′ genes (Fig. 4.6). Mutations of Hox genes result in morphological transformations of the segmental structures in which a specific gene is normally expressed. Generally, loss-of-function mutations result in posterior-to-anterior transformations (e.g., cells of a given segment form the structural equivalent of the next most anterior segment), and gain-of-function mutations result in anterior-to-posterior structural transformations. Figure 4.7 illustrates an experiment in which injection of an antibody to a homeodomain protein into an early frog embryo resulted in the transformation of the anterior spinal cord into an expanded hindbrain.
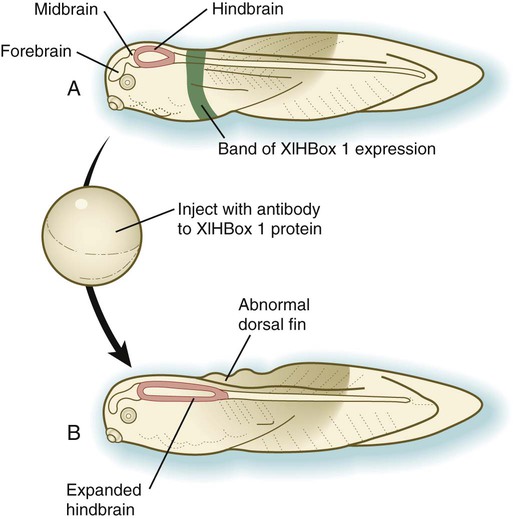
A, Normal larva, showing a discrete band (green) of XlHbox 1 expression. B, Caudal expansion of the hindbrain after antibodies to XlHbox 1 protein are injected into the early embryo. (Based on Wright CV and others: Cell 59:81-93, 1989.)
Although Hox genes were originally described to operate along the main body axis, sequential arrays of expression are found in developing organs or regions as diverse as the gut, the limbs, and the internal and external genitalia. The expression of isolated Hox genes also occurs in locations such as hair follicles, blood cells, and developing sperm cells. The principal function of the Hox genes is involved in setting up structures along the main body axis, but ordered groups of Hox genes are later reused in guiding the formation of several specific nonaxial structures. In mammals, individual members of a paralogous group often have similar functions, so that if one Hox gene is inactivated, the others of that paralogous group may compensate for it. If all members of a paralogous group are inactivated, profound morphological disturbances often result (see p. 171).
Pax Genes
The Pax gene family, consisting of 9 known members, is an important group of genes that are involved in many aspects of mammalian development (Fig. 4.8). The Pax genes are homologous to the Drosophila pair-rule segmentation genes (see Fig. 4.1). All Pax proteins contain a paired domain of 128 amino acids that binds to DNA. Various members of this group also contain entire or partial homeobox domains and a conserved octapeptide sequence. Pax genes play a variety of important roles in the sense organs and developing nervous system, and outside the nervous system they are involved in cellular differentiative processes when epithelial-mesenchymal transitions occur.
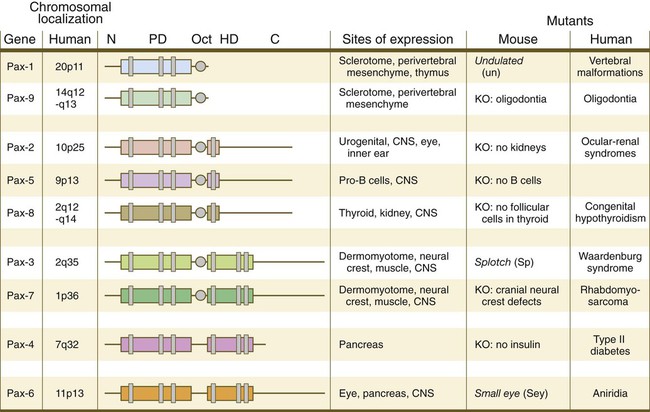
The structures of conserved elements of these genes are schematically represented. CNS, central nervous system; KO, knockout. (Modified from Wehr R, Gruss P: Int J Dev Biol 40:369-377, 1996; and Epstein JC: Trends Cardiovasc Med 6:255-260, 1996.)
Other Homeobox-Containing Gene Families
The POU gene family is named for the acronym of the first genes identified: Pit1, a gene uniquely expressed in the pituitary; Oct1 and Oct2; and Unc86, a gene expressed in a nematode. Genes of the POU family contain, in addition to a homeobox, a region encoding 75 amino acids, which also bind to DNA through a helix-loop-helix structure. As described in Chapter 3 (see p. 42), Oct-4 plays an important role during early cleavage.
The Lim proteins constitute a large family of homeodomain proteins, some of which bind to the DNA in the nucleus and others of which are localized in the cytoplasm. Lim proteins are involved at some stage in the formation of virtually all parts of the body. The absence of certain Lim proteins results in the development of headless mammalian embryos (see p. 83).
Helix-Loop-Helix Transcription Factors
Basic Helix-Loop-Helix Proteins
The transcription factors of the basic helix-loop-helix type are proteins that contain a short stretch of amino acids in which two α-helices are separated by an amino acid loop. This region, with an adjacent basic region, allows the regulatory protein to bind to specific DNA sequences. The basic regions of these proteins bind to DNA, and the helix-loop-helix domain is involved in homodimerization or heterodimerization. This configuration is common in numerous transcription factors that regulate myogenesis (see Fig. 9.33).
Zinc Finger Transcription Factors
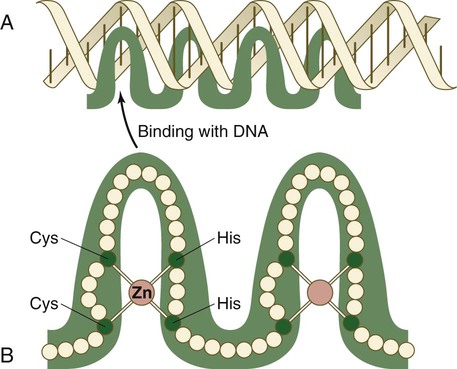
The zinc finger family of transcription factors consists of proteins with regularly placed cystidine and histidine units that are bound by zinc ions to cause the polypeptide chain to pucker into fingerlike structures (Fig. 4.9). These “fingers” can be inserted into specific regions in the DNA helix.
Sox Genes
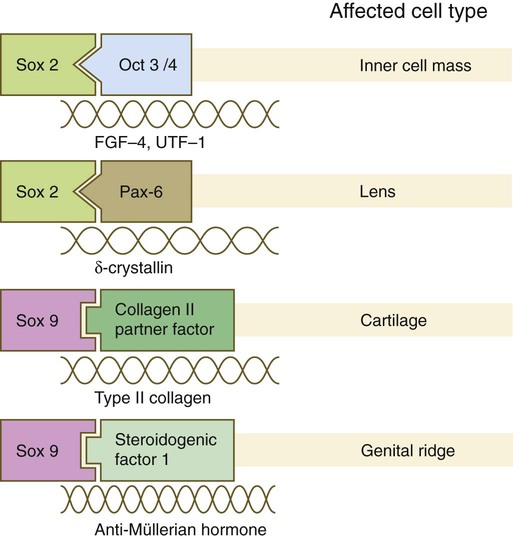
The Sox genes comprise a large family (>20 members) that have in common an HMG (high-mobility group) domain on the protein. This domain is unusual for a transcription factor in that, with a partner protein, it binds to 7 nucleotides on the minor instead of the major groove on the DNA helix and causes a pronounced conformational change in the DNA. Sox proteins were first recognized in 1990, when the SRY gene was shown to be the male-determining factor in sex differentiation (see p. 389), and the name of this group, Sox, was derived from Sry HMG box. One characteristic of Sox proteins is that they work in concert with other transcription factors to influence expression of their target genes (Fig. 4.10). As may be expected from their large number, Sox proteins are expressed by most structures at some stage in their development.
Signaling Molecules
Transforming Growth Factor-β Family
The transforming growth factor- β (TGF-β) superfamily consists of numerous molecules that play a wide variety of roles during embryogenesis and postnatal life. The TGF family was named because its first-discovered member (TGF-β1) was isolated from virally transformed cells. Only later was it realized that many signaling molecules with greatly different functions during embryonic and postnatal life bear structural similarity to this molecule. Table 4.1 summarizes some of these molecules and their functions.
Table 4.1
Members of the Transforming Growth Factor-β Superfamily Mentioned in This Text
| Member | Representative Functions | Chapters |
| TGF-β1 to TGF-β5 | Mesodermal induction | 5 |
| Myoblast proliferation | 9 | |
| Invasion of cardiac jelly by atrioventricular endothelial cells | 17 | |
| Activin | Granulosa cell proliferation | 1 |
| Mesodermal induction | 5 | |
| Inhibin | Inhibition of gonadotropin secretion by hypophysis | 1 |
| Müllerian inhibiting substance | Regression of paramesonephric ducts | 16 |
| Decapentaplegic | Signaling in limb development | 10 |
| Vg1 | Mesodermal and primitive streak induction | 5 |
| BMP-1 to BMP-15 | Induction of neural plate, induction of skeletal differentiation, and other inductions | 5, 9, 10 |
| Nodal | Formation of mesoderm and primitive streak, left-right axial fixation | 5 |
| Glial cell line–derived neurotrophic factor | Induction of outgrowth of ureteric bud, neural colonization of gut | 16, 12 |
| Lefty | Determination of body asymmetry | 5 |
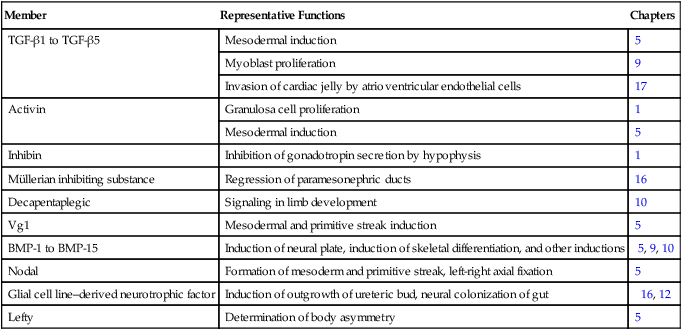
BMP, bone morphogenetic protein; TGF-β, transforming growth factor-β.
The formation, structure, and modifications of TGF-β1 are representative of many types of signaling molecules and are used as an example (Fig. 4.11). Similar to many members of this family, TGF-β1 is a disulfide-linked dimer, which is synthesized as a pair of inactive 390-amino acid precursors. The glycosylated precursor consists of a small N-terminal signal sequence, a much larger proregion, and a 112-amino acid C-terminal bioactive domain. The proregion is enzymatically cleaved off the bioactive domain at a site of 4 basic amino acids adjoining the bioactive domain. After secretion from the cell, the proregion of the molecule remains associated with the bioactive region, thus causing the molecule to remain in a latent form. Only after dissociation of the proregion from the bioactive region does the bioactive dimer acquire its biological activity.
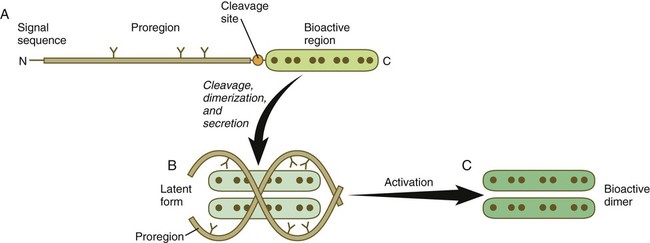
A, The newly synthesized peptide consists of a C-terminal bioactive region, to which is attached a long glycosylated proregion and an N-terminal signal sequence. B, The proregion is cleaved off from the bioactive region, and two secreted bioactive regions form a dimer that is maintained in a latent form by being complexed with the separated proregions. C, Through an activation step, the bioactive dimer is released from the proregions and can function as a signaling molecule.
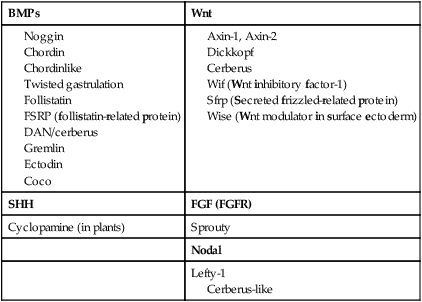
Among the most important subfamilies of the TGF-β family are the bone morphogenetic proteins (BMPs). Although BMP was originally discovered to be the active agent in the induction of bone during fracture healing, the 15 members of this group play important roles in the development of most structures in the embryo. BMPs often exert their effects by inhibiting other processes in the embryo. To make things even more complicated, certain very important interactions in embryonic development (e.g., induction of the central nervous system; see p. 84) occur because of the inhibition of BMP by some other molecule. The net result is an effect caused by the inhibition of an inhibitor. Molecules that inhibit or antagonize the action of BMPs are listed in Table 4.2. These molecules bind to secreted BMP dimers and interfere with their binding to specific receptors.
Cerberus-like
Fibroblast Growth Factor Family
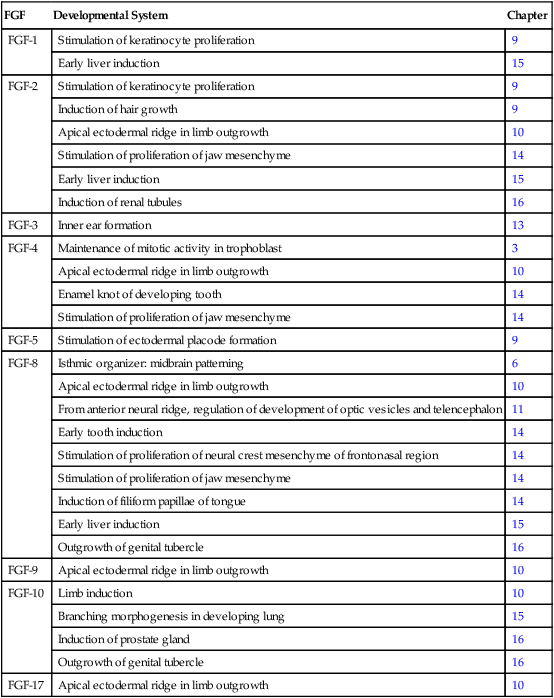
Fibroblast growth factor (FGF) was initially described in 1974 as a substance that stimulates the growth of fibroblasts in culture. Since then, the originally described FGF has expanded into a family of 22 members, each of which has distinctive functions. Many members of the FGF family play important roles in a variety of phases of embryonic development and in fulfilling functions, such as the stimulation of capillary growth, in the postnatal body. Some of the functions of the FGFs in embryonic development are listed in Table 4.3. Secreted FGFs are closely associated with the extracellular matrix and must bind to heparan sulfate to activate their receptors.
Table 4.3
Members of the Fibroblast Growth Factor Family Mentioned in This Text
| FGF | Developmental System | Chapter |
| FGF-1 | Stimulation of keratinocyte proliferation | 9 |
| Early liver induction | 15 | |
| FGF-2 | Stimulation of keratinocyte proliferation | 9 |
| Induction of hair growth | 9 | |
| Apical ectodermal ridge in limb outgrowth | 10 | |
| Stimulation of proliferation of jaw mesenchyme | 14 | |
| Early liver induction | 15 | |
| Induction of renal tubules | 16 | |
| FGF-3 | Inner ear formation | 13 |
| FGF-4 | Maintenance of mitotic activity in trophoblast | 3 |
| Apical ectodermal ridge in limb outgrowth | 10 | |
| Enamel knot of developing tooth | 14 | |
| Stimulation of proliferation of jaw mesenchyme | 14 | |
| FGF-5 | Stimulation of ectodermal placode formation | 9 |
| FGF-8 | Isthmic organizer: midbrain patterning | 6 |
| Apical ectodermal ridge in limb outgrowth | 10 | |
| From anterior neural ridge, regulation of development of optic vesicles and telencephalon | 11 | |
| Early tooth induction | 14 | |
| Stimulation of proliferation of neural crest mesenchyme of frontonasal region | 14 | |
| Stimulation of proliferation of jaw mesenchyme | 14 | |
| Induction of filiform papillae of tongue | 14 | |
| Early liver induction | 15 | |
| Outgrowth of genital tubercle | 16 | |
| FGF-9 | Apical ectodermal ridge in limb outgrowth | 10 |
| FGF-10 | Limb induction | 10 |
| Branching morphogenesis in developing lung | 15 | |
| Induction of prostate gland | 16 | |
| Outgrowth of genital tubercle | 16 | |
| FGF-17 | Apical ectodermal ridge in limb outgrowth | 10 |
Hedgehog Family
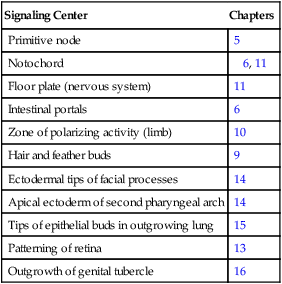
The hedgehog signaling molecules burst on the vertebrate embryological scene in 1994 and are among the most important signaling molecules known (Table 4.4). Related to the segment-polarity molecule, hedgehog, in Drosophila, the three mammalian hedgehogs have been given the whimsical names of desert, Indian, and sonic hedgehog. The name hedgehog arose because mutant larvae in Drosophila contain thick bands of spikey outgrowths on their bodies.
Table 4.4
Sites in the Embryo Where Sonic Hedgehog Serves as a Signaling Molecule
| Signaling Center | Chapters |
| Primitive node | 5 |
| Notochord | 6, 11 |
| Floor plate (nervous system) | 11 |
| Intestinal portals | 6 |
| Zone of polarizing activity (limb) | 10 |
| Hair and feather buds | 9 |
| Ectodermal tips of facial processes | 14 |
| Apical ectoderm of second pharyngeal arch | 14 |
| Tips of epithelial buds in outgrowing lung | 15 |
| Patterning of retina | 13 |
| Outgrowth of genital tubercle | 16 |
Sonic hedgehog (shh) is a protein with a highly conserved N-terminal region and a more divergent C-terminal region. After its synthesis and release of the propeptide from the rough endoplasmic reticulum, the signal peptide is cleaved off, and glycosylation occurs on the remaining peptide (Fig. 4.12). Still within the cell, the shh peptide undergoes autocleavage through the catalytic activity of its C-terminal portion. During cleavage, the N-terminal segment becomes covalently bonded with cholesterol. The 19-kD N-terminal peptide is secreted from the cell, but it remains bound to the surface of the cell that produced it. All the signaling activity of shh resides in the N-terminal segment. Through the activity of another gene product (disp [dispatched] in Drosophila), the N-terminal segment of shh, still bound with cholesterol, is released from the cell. The C-terminal peptide plays no role in signaling.
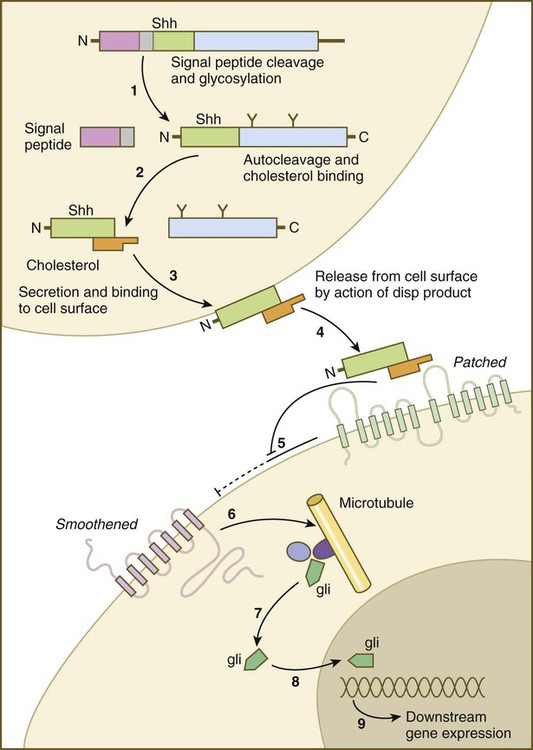
(1) The signal peptide is cleaved off the newly synthesized polypeptide, and the remainder undergoes glycosylation; (2) the remaining peptide undergoes autocleavage under the influence of the C-terminal portion, and cholesterol binds to the N-terminal part, which is the active part of the molecule; (3) the N-terminal part is secreted and bound to the cell surface; (4) the bound shh molecule is released from the cell surface through the action of a product of dispersed (disp); (5) the released shh inhibits the inhibitory effect of Patched on smoothened; (6) on release from the inhibitory influence of Patched, smoothened emits a signal that (7) releases the transcription factor Gli from a complex of molecules bound to microtubules; (8) Gli enters the nucleus and binds to the DNA, (9) influencing the expression of many genes.
Wnt Family
Wnts have been described as being “stickier” than other signaling molecules, and they often interact with components of the extracellular matrix. Their signaling pathway is complex and is still not completely understood (see Fig. 4.16). Similar to most other signaling molecules, the activity of Wnts can be regulated by other inhibitory molecules (see Table 4.2). Some inhibitory molecules, such as Wnt-inhibitory factor-1 (WIF-1) and cerberus, directly bind to the Wnt molecule. Others, such as dickkopf, effect inhibition by binding to the receptor complex.
Other Actions of Signaling Molecules
An important and more recent realization in molecular embryology is how often signaling molecules act by inhibiting the actions of other signaling molecules. For example, the signaling molecules chordin, noggin, and gremlin all inhibit the activity of BMP, which itself often acts as an inhibitor (see Table 4.2).
Receptor Molecules
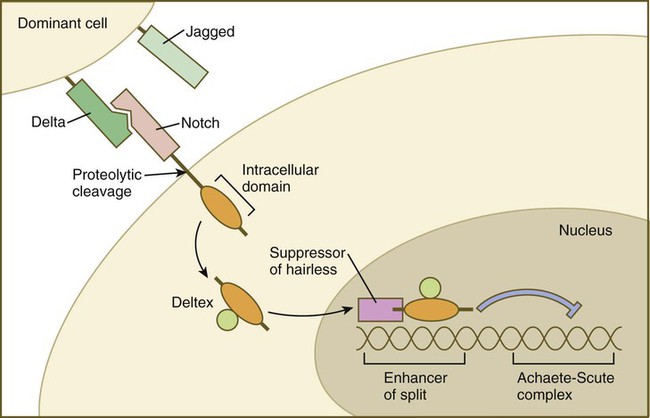
Cell surface receptors are typically transmembrane proteins with extracellular, transmembrane, and cytoplasmic domains (see Fig. 4.2). The extracellular domain contains a binding site for the ligand, which is typically a hormone, cytokine, or growth factor. When the ligand binds to a receptor, it effects a conformational change in the cytoplasmic domain of the receptor molecule. Cell surface receptors are of two main types: (1) receptors with intrinsic protein kinase activity and (2) receptors that use a second messenger system to activate cytoplasmic protein kinases. An example of the first type is the family of receptors for FGFs, in which the cytoplasmic domain possesses tyrosine kinase activity. Receptors for growth factors of the TGF-β superfamily are also of this type, but in them the cytoplasmic domain contains serine/threonine kinase activity. In cell surface receptors of the second type, the protein kinase activity is separate from the receptor molecule itself. This type of receptor is also activated by binding with a ligand (e.g., neurotransmitter, peptide hormone, growth factor), but a series of intermediate steps is required to activate cytoplasmic protein kinases. A surface receptor, Notch, is introduced in greater detail in Box 4.2 as a specific example of a receptor that plays many important roles in embryonic development.
Signal Transduction
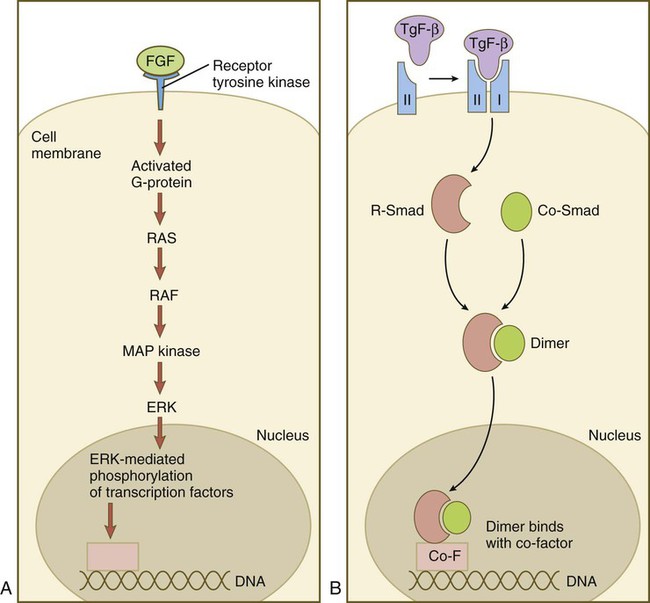
Members of the FGF family connect with the receptor tyrosine kinase (TRK) pathway (Fig. 4.15A). After FGF has bound to the receptor, a G protein near the receptor becomes activated and sets off a long string of intracytoplasmic reactions, starting with RAS and ending with the entry of ERK into the nucleus, and its interaction with transcription factors. Members of the TGF-β family first bind to a type II serine/threonine kinase receptor, which complexes with a type I receptor (Fig. 4.15B). This process activates a pathway dominated by Smad proteins. Two different Smads (R-Smad and Co-Smad) dimerize and enter the nucleus. The Smad dimer binds with a cofactor and is then capable of binding with some regulatory element on the DNA.
The hedgehog pathway was already introduced in Figure 4.12. The complex Wnt pathway first involves binding of the Wnt molecule to its transmembrane receptor, Frizzled. In a manner not yet completely understood, Frizzled interacts with the cytoplasmic protein Disheveled, which ties up a complex of numerous molecules (destruction complex), which in the absence of Wnt cause the degradation of an important cytoplasmic protein, β-catenin (Fig. 4.16). If β-catenin is not destroyed, it enters the nucleus, where it acts as a powerful adjunct to transcription factors that determine patterns of gene expression.
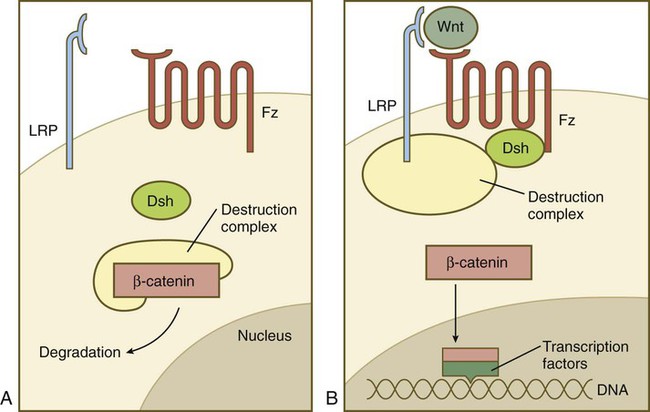
A, In the absence of a Wnt signal, β-catenin is bound in a destruction complex and is degraded. B, In the presence of Wnt, the receptor Frizzled (Fz) activates Disheveled (Dsh), which prevents the destruction complex from degrading β-catenin. β-catenin then enters the nucleus, where it forms complexes with transcription factors.
Small RNAs
Although small RNAs function through a bewildering array of mechanisms, one major pathway is close to being common (Fig. 4.17). miRNAs often begin as double-stranded molecules with a hairpin loop. Through the activity of an enzyme called Dicer, the miRNA precursor is cleaved, resulting in a single-stranded miRNA, which is then bound to a member of the Argonaute (AGO) protein family. In many cases, the AGO-siRNA complex has RNase activity and is able to disrupt a target RNA molecule enzymatically. In this way specific gene expression is modulated. By applying this principle, developmental geneticists are able to target the disruption of specific genes under investigation by interfering with the mRNAs that these genes produce.
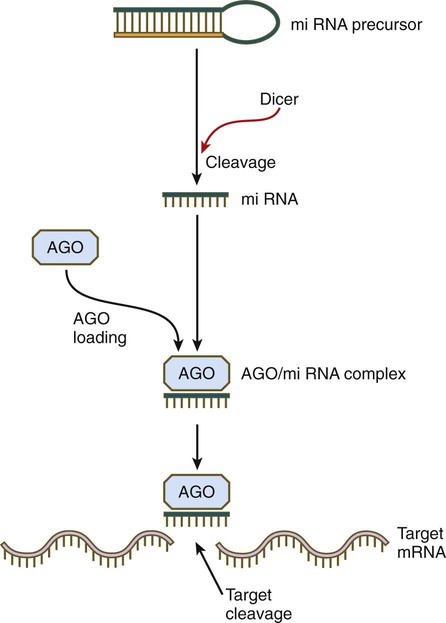
The double helical precursor molecule, often containing a hairpin loop, is cleaved by Dicer, resulting in a small miRNA molecule, which is then complexed with an Argonaute (AGO) protein. This complex approaches the target mRNA and through its intrinsic RNase activity, it cleaves the target mRNA molecule, thereby inactivating it.
Retinoic Acid
Vitamin A enters the body of the embryo as retinol and binds to a retinol-binding protein, which attaches to specific cell surface receptors (Fig. 4.18). Retinol is released from this complex and enters the cytoplasm, where it is bound to cellular retinol-binding protein (CRBP I). In the cytoplasm, the all-trans retinol is enzymatically converted first to all-trans retinaldehyde and then to all-trans retinoic acid, the retinoid with the most potent biological activity (see Fig. 4.18). CRBP and CRABP I (cellular retinoic acid–binding protein) may function to control the amount of retinoids that enters the nucleus. When released from CRABP, retinoic acid enters the nucleus, where it typically binds to a heterodimer consisting of a member of the retinoic acid receptor (RAR) α, β, or γ family and a member of the retinoid X receptor (RXR) α, β, or γ family. This complex of retinoic acid and receptor heterodimer binds to a retinoic acid response element (RARE) on DNA, usually on the enhancer region of a gene, and it acts as a transcription factor, controlling the production of a gene product.
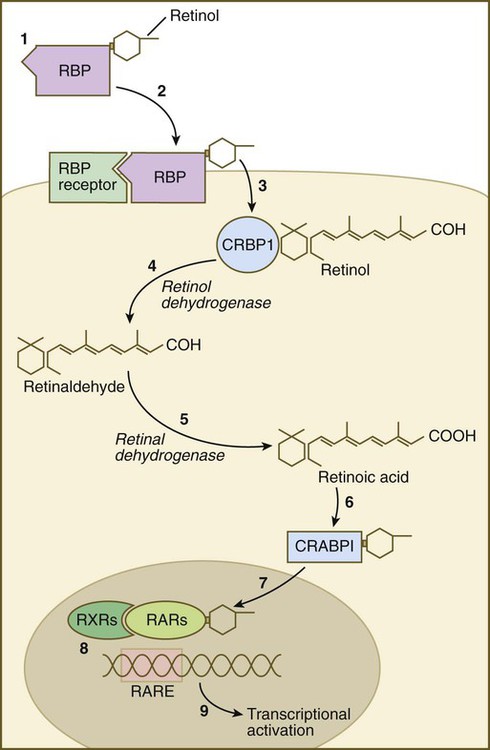
(1) Retinol becomes bound to a retinol-binding protein (RBP) outside the cell; (2) this complex is bound to an RBP receptor on the cell surface; (3) the retinol is released into the cytoplasm and is bound to a cytoplasmic RBP (CRBP I); (4) through the action of retinol dehydrogenase, retinol is converted to retinaldehyde (5), which is converted to retinoic acid by retinal dehydrogenase; (6) retinoic acid is bound to a cytoplasmic receptor (CRABP I) and taken into the nucleus; (7) within the nucleus, retinoic acid is bound to a dimer of two nuclear retinoic acid receptors (RXR and RAR); (8) this complex binds to a retinoic acid response element (RARE) on the DNA and (9) activates transcription of target genes.
Retinoic acid is produced and used in specific local regions at various times during prenatal and postnatal life. Among its well-defined targets early in development are certain Hox genes (e.g., Hoxb-1); misexpression of these genes caused by either too little or too much retinoic acid can result in serious disturbances in the organization of the hindbrain and pharyngeal neural crest. One of the most spectacular examples of the power of retinoic acid is its ability to cause extra pairs of limbs to form alongside the regenerating tails of amphibians (Fig. 4.19). This is a true example of a homeotic shift in a vertebrate, similar to the formation of double-winged flies or legs instead of antennae in Drosophila (see p. 59).
Summary
 Evidence is increasing that the basic body plan of mammalian embryos is under the control of many of the same genes that have been identified as controlling morphogenesis in Drosophila. In this species, the basic axes are fixed through the actions of maternal-effect genes. Batteries of segmentation genes (gap, pair-rule, and segment-polarity genes) are then activated. Two clusters of homeotic genes next confer a specific morphogenetic character to each body segment. Because of their regulative nature, mammalian embryos are not as rigidly controlled by genetic instructions as are Drosophila embryos.
Evidence is increasing that the basic body plan of mammalian embryos is under the control of many of the same genes that have been identified as controlling morphogenesis in Drosophila. In this species, the basic axes are fixed through the actions of maternal-effect genes. Batteries of segmentation genes (gap, pair-rule, and segment-polarity genes) are then activated. Two clusters of homeotic genes next confer a specific morphogenetic character to each body segment. Because of their regulative nature, mammalian embryos are not as rigidly controlled by genetic instructions as are Drosophila embryos.
 The homeobox, a highly conserved region of 180 base pairs, is found in multiple different genes in almost all animals. The homeobox protein is a transcription factor. Homeobox-containing genes are arranged along the chromosome in a specific order and are expressed along the craniocaudal axis of the embryo in the same order. Activation of homeobox genes may involve interactions with other morphogenetically active agents, such as retinoic acid and TGF-β.
The homeobox, a highly conserved region of 180 base pairs, is found in multiple different genes in almost all animals. The homeobox protein is a transcription factor. Homeobox-containing genes are arranged along the chromosome in a specific order and are expressed along the craniocaudal axis of the embryo in the same order. Activation of homeobox genes may involve interactions with other morphogenetically active agents, such as retinoic acid and TGF-β.
 Many of the molecules that control development can be assigned to several broad groups. One group is the transcription factors, of which the products of homeobox-containing genes are just one of many types. A second category is signaling molecules, many of which are effectors of inductive interactions. Some of these are members of large families, such as the TGF-β and FGF families. An important class of signaling molecules is the hedgehog proteins, which mediate the activities of many important organizing centers in the early embryo. Signaling molecules interact with responding cells by binding to specific surface or cytoplasmic receptors. These receptors represent the initial elements of complex signal transduction pathways, which translate the signal to an intracellular event that results in new patterns of gene expression in the responding cells. Small RNAs play important roles in the control of gene expression, mainly at posttranscriptional levels. Retinoic acid (vitamin A) is a powerful, but poorly understood, developmental molecule. Misexpression of retinoic acid causes level shifts in axial structures through interactions with Hox genes.
Many of the molecules that control development can be assigned to several broad groups. One group is the transcription factors, of which the products of homeobox-containing genes are just one of many types. A second category is signaling molecules, many of which are effectors of inductive interactions. Some of these are members of large families, such as the TGF-β and FGF families. An important class of signaling molecules is the hedgehog proteins, which mediate the activities of many important organizing centers in the early embryo. Signaling molecules interact with responding cells by binding to specific surface or cytoplasmic receptors. These receptors represent the initial elements of complex signal transduction pathways, which translate the signal to an intracellular event that results in new patterns of gene expression in the responding cells. Small RNAs play important roles in the control of gene expression, mainly at posttranscriptional levels. Retinoic acid (vitamin A) is a powerful, but poorly understood, developmental molecule. Misexpression of retinoic acid causes level shifts in axial structures through interactions with Hox genes.
 Many cancers are caused by mutations of genes involved in normal development. Two major classes of cancer-causing genes are proto-oncogenes, which induce tumor formation through gain-of-function mechanisms, and tumor suppressor genes, which cause cancers through loss-of-function mutations.
Many cancers are caused by mutations of genes involved in normal development. Two major classes of cancer-causing genes are proto-oncogenes, which induce tumor formation through gain-of-function mechanisms, and tumor suppressor genes, which cause cancers through loss-of-function mutations.