CHAPTER 9 Membrane Carriers
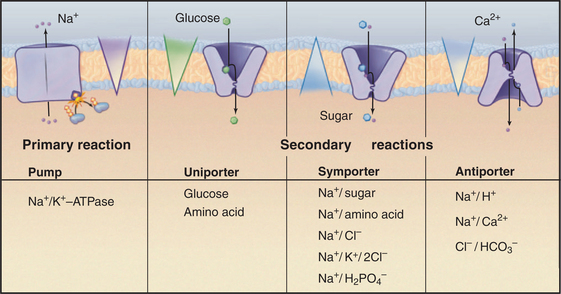
Carriers are integral membrane proteins that use electrochemical gradients to move select chemical substrates across lipid bilayers (Fig. 9-1). Transport by well-characterized carriers depends on a conformational change to move each substrate. Typically, the carriers work step by step, more like enzymes than channels. Channels simply provide a selective pore for transport, and they generally transport at much higher rates (see Chapter 10). Common substrates for carriers are ions and small soluble organic molecules, but in some cases, substrates are lipid soluble.
When a carrier uses an ion gradient to provide the energy to transport a substrate, it is said to catalyze a secondary reaction. In this sense, pumps catalyze primary transport reactions, using energy from ATP hydrolysis, electron transport, or absorption of light to create ion gradients (see Chapter 8). Coupling an ion gradient created by pumps to drive transport by a carrier is called a chemiosmotic cycle (see Fig. 11-1).
Diversity of Carrier Proteins
Biological experimentation and exploration of genomes have revealed more than a hundred families of carriers, many of which can be grouped into superfamilies. The major facilitator superfamily (MFS) is the focus of this chapter, since it includes about one third of all known carrier proteins, including many of the best-characterized carriers. Thousands of MFS genes in all branches of the phylogenetic tree are likely to have arisen from a common ancestor. Two thirds of known carriers have other origins and structures but have converged on mechanical solutions for solute transport similar to MFS carriers. No one knows how many structurally distinct groups exist in nature. Box 9-1 illustrates a small selection of carrier families with different evolutionary origins and structures.
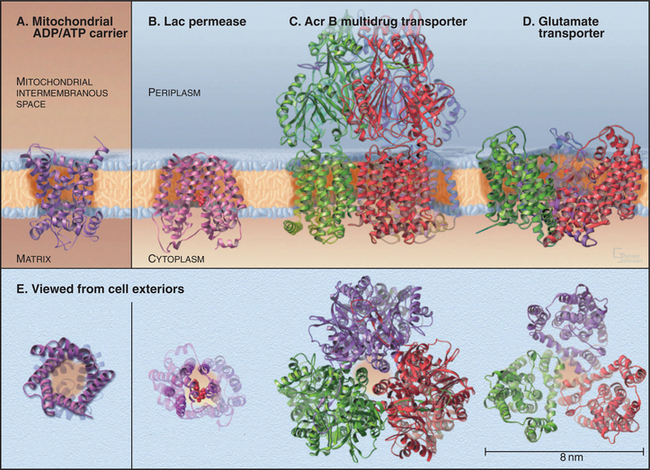
BOX 9-1 Crystal Structures of Diverse Carrier Proteins
Four crystal structures (Fig. 9-2) illustrate the diversity of carrier proteins. They differ in evolutionary origins and structures, but all function as carriers. They converged toward common mechanisms implemented by different structures. Conformational changes are believed to contribute to transport in all cases, so their mechanisms will be better understood when structures of additional conformations of each protein are available.
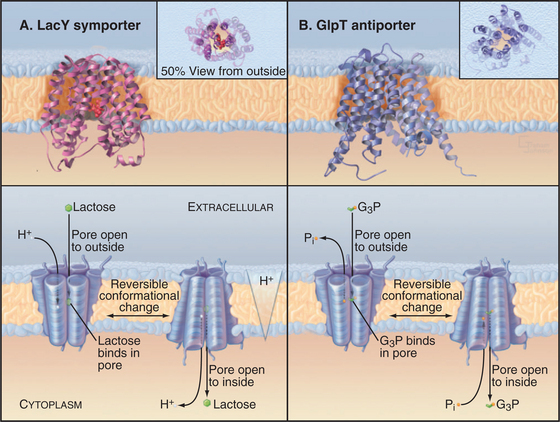
Structure of MFS Carrier Proteins
Crystal structures of two MFS carrier proteins from Escherichia coli (Fig. 9-3A-B) confirmed much of what had been learned about their organization from less direct methods. GlpT is a glycerol-3-phosphate–phosphate antiporter. LacY, the lactose permease, is a lactose-proton symporter. Both proteins consist of 12 transmembrane α-helices. The sequences and structures of the two halves of each protein are homologous, so it is believed that the original gene was created by duplication of an ancestral gene, which coded for a six-helix protein that formed functional dimers. As the MFS gene family grew during evolution, the ancient gene duplication and fusion process had two advantages. First, it allowed the two halves of each gene to diversify separately to increase specificity for a wide variety of substrates. Second, a single polypeptide simplifies assembly of a functional carrier, as two half-sized subunits do not have to find each other. If the two halves of a 12-helix MFS carrier are expressed in the same cell, they can assemble functional carriers, but less efficiently than the intact protein.
Carrier Physiology and Mechanisms
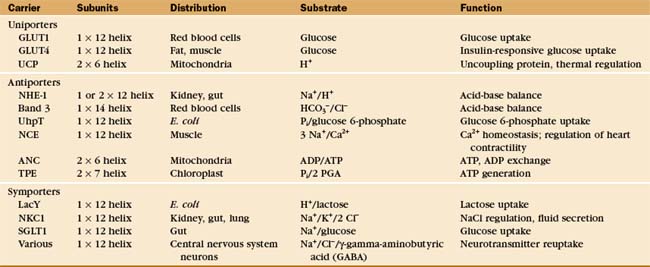
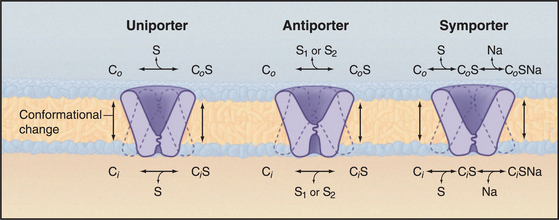
Investigators have identified about 500 different reactions that are attributable to secondary transporters and have characterized about a dozen carriers well enough to understand their mechanisms. These model systems (Table 9-1) provide a framework to divide carriers into three broad classes (Fig. 9-4) based on mechanism:
A few carriers are more complicated than is indicated by this classification. For example, neurotransmitter carriers catalyze both antiporter and symporter reactions, with Na+ and Cl− going in one direction and a neurotransmitter in the opposite direction. This example also makes the important general point that the stoichiometry of antiporter and symporter reactions need not be one-to-one. Table 9-1 provides other examples.
All three classes of MFS carriers use similar mechanisms to transfer bound substrates across membranes. This follows naturally from having a common evolutionary ancestor and similar architectures. They work like enzymes, binding substrates on one side of membranes, undergoing a conformational change that reorients this binding site, and releasing substrate on the opposite side of the membrane. Substrate concentrations on the two sides determine the direction of net transfer across the membrane. Whether a carrier is a uniporter, antiporter, or symporter depends simply on the number of substrate-binding sites and the rate and equilibrium constants for the various species to reorient across the membrane. The actual rate of transfer depends on the concentrations of substrates. A limited number of specific inhibitors (Table 9-2) have been useful in establishing physiological functions of carriers.
| Agent | Target |
|---|---|
| Furosemide* | Na+/K+/2 Cl− symporter |
| Amiloride* | Na+/H+ antiporter |
| SITZ, DITZ | HCO3− /Cl− antiporter |
| Cytochalasin B | GLUT isoforms |
| Phloretin | GLUT isoforms |
| Phlorizin | SGLT isoforms |
Uniporters
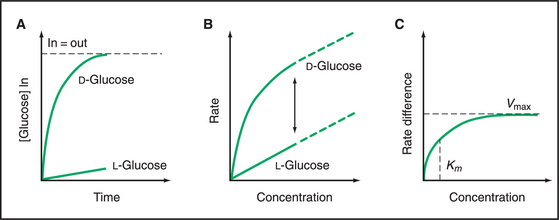
Classic experiments with GLUT1 in the plasma membrane of red blood cells led to the carrier concept in the 1950s. Human red blood cells are convenient to use because they express high concentrations of GLUT carriers and because blood banks can provide large quantities of cells. The time course of radioactive glucose accumulation (Fig. 9-5A) shows that transport is stereospecific for δ−glucose and that net transport stops when the concentrations of δ−glucose are equal inside and out. Slow equilibration of l-glucose across the membrane probably represents passive diffusion across the lipid bilayer, as this rate can be predicted from the solubility of glucose in membrane lipids. This experiment showed that something in the membrane causes an acceleration in the rate of glucose entry, giving rise to the concept of facilitated diffusion.
The dependence of the initial rate of δ−glucose entry on its concentration (Fig. 9-5B) provided evidence for a specific, saturable, carrier molecule in the membrane. Because the rate of δ−glucose entry includes both diffusion across the bilayer and movement through a carrier, the l-glucose rate must be used to correct for the rate of diffusion. Once this has been done, the rate of facilitated δ−glucose entry has a hyperbolic dependence on the concentration of δ−glucose (Fig. 9-5C). This concentration dependence is just like a bimolecular binding reaction (see Fig. 4-2) or the rate of a simple enzyme mechanism that depends on the rate of substrate binding to the enzyme. Thus, the substrate concentration at the half-maximal velocity provides an estimate of the affinity of the carrier for the substrate. At high substrate concentrations, the substrate-binding sites on the carrier are saturated, and the rate plateaus at a maximal velocity owing to rate-limiting conformational changes. These enzyme-like properties, along with the ability to stop facilitated transport with protein inhibitors, suggest that carriers are proteins with specific binding sites for their substrates.
The carrier hypothesis is now understood in terms of carrier proteins embedded in the lipid bilayer; these proteins bind substrate and undergo readily reversible first-order transitions between at least two different conformations (Fig. 9-3). One conformation exposes a substrate-binding site on one side of the membrane. Another conformation exposes a binding site on the other side. Thus, a single substrate molecule can bind on one side of the membrane and be released on the other, thus moving across the membrane. (The carrier does not physically diffuse across the membrane, as was formerly believed.)
Net transport requires a concentration gradient, as the conformational change that reorients substrate-binding sites is reversible, and substrate can move either way. The rates of substrate movement depend on the rates of formation of substrate-carrier complex on the two sides of the membrane. The rates of these second-order reactions depend directly on the substrate concentrations, so the binding sites of the carrier are more fully occupied on the uphill side, and the net movement of substrate is therefore in the downhill direction. When substrate concentrations are the same on the two sides, exchange continues without net movement because the carrier is equally saturated on both sides of the membrane.
This simple carrier model clarified a large body of confusing information and clearly distinguished car-riers from channels for the first time. The original car-rier model for the GLUT1 uniporter also led directly to kinetic schemes for antiporters and symporters (Fig. 9-4).
See Figure 11-2 for a depiction of epithelial cells using glucose symporters and uniporters to take up glucose from the intestine after a meal. See Figure 28-6 for an illustration of brown fat cells using uncoupling protein, a proton uniporter in mitochondria, to generate heat when animals arise from hibernation and mammalian infants are born.
Antiporters
Antiporters translocate two different substrates and use a concentration gradient of one substrate to drive another substrate up its concentration gradient. Like uniporters, these carriers undergo reversible, conformational changes that expose substrate-binding sites on one or the other side of a membrane. Two modifications of the uniporter mechanism provide a model for antiporters (Fig. 9-4). First, two substrates, S1 and S2, compete for binding antiporters. Second, a substrate-free carrier cannot undergo the conformational changes required to change the orientation of its binding sites. These differences make transport of two substrates dependent on each other in an obligate fashion.
For example, the heart 3Na+/Ca2+ exchanger binds either Na+ or Ca2+ and uses the large Na+ gradient across the plasma membrane to drive the transport of Ca2+ out of the cytoplasm up a concentration gradient (see Fig. 11-13). On the outer surface of the cell, the carrier binds three Na+. After the conformational change that reorients the binding site, the three Na+ dissociate inside and one Ca2+ binds. Reorientation of the binding site carries this Ca2+ to the outer surface of the cell, where it dissociates. (In addition to the substrate concentrations, the membrane potential must also be taken into account, as this exchange is not electrically neutral, and the potential may affect the binding of one or both substrates to the carrier.)
Symporters
The prefix “sym-” indicates that two substrates are transported in the same direction. A simple extension of the uniporter mechanism provides a model for symporters (Fig. 9-4). Like uniporters, the carrier has two conformations with substrate-binding sites open to either side of the membrane. The binding site may be free or occupied by a single substrate, like a uniporter, but in addition, two substrates can bind together. A second difference is that transmembrane reorientation of substrate-binding sites is much more favorable for free carrier and carrier with two bound substrates than for carrier with only one bound substrate. This feature of symporters minimizes leaks of one substrate across the membrane.
E. coli LacY symporter uses a proton gradient across the plasma membrane to drive accumulation of lactose (Fig. 9-3A). The proton gradient is created by the respiratory chain under aerobic conditions or by the F-type ATPase pump under anaerobic conditions (see Fig. 8-5). Protons move down their concentration gradient as lactose moves up its concentration gradient into the cell. Mutations in LacY can cause internal leaks that uncouple sugar transport from proton movements. Vertebrate SGLT1 symporter carries out a comparable reaction for intestinal epithelial cells using the Na+ gradient to take up glucose from the lumen of the gut (see Fig. 11-2).
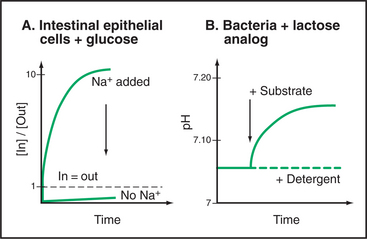
Two key experiments (Fig. 9-6) established the symporter concept. The first demonstrated that extracellular Na+ was required for intestinal cells with the SGLT1 transporter to accumulate δ−glucose against a concentration gradient. This experiment left open the possibility that Na+ simply activates the carrier in some way without being used directly to drive glucose accumulation. Second, an experiment with LacY demonstrated that sugar and cation move across the membrane together. Not only was a proton gradient required for sugar transport but also a high concentration of another sugar (a nonmetabolizable lactose analog) in the medium could drive H+ into the cell along with the sugar. Additional experiments confirmed that the stoichiometry of the reaction was one lactose transported in for every H+ transported in. (Because this transport reaction is not electrically neutral, the membrane potential is a factor, and another pathway must be available to balance the charge—e.g., by carrying k+ in the opposite direction.) These experiments established that both substrates move together across the membrane with a fixed stoichiometry and that a concentration gradient of either can drive the other. Parallel experiments on plasma membrane vesicles isolated from vertebrate kidney cells showed that Na+ moving inward down its electrochemical gradient can drag glucose in with it. When the Na+ concentration is equal on the two sides, the carrier facilitates the movement of glucose across the membrane but not its net accumulation.
Abramson J, Kaback HR, Iwata S. Structural comparison of lactose permease and the glycerol-3-phosphate antiporter: Members of the major facilitator superfamily. Curr Opin Struct Biol. 2004;14:413-419.
Abramson J, Smirnova I, Kasho V, et al. Structure and mechanism of the lactose permease of E. coli. Science. 2003;301:610-615.
Guan L, Kaback HR. Lessons from lactose permease. Annu Rev Biophys Biomol Struct. 2006;35:67-91.
Huang Y, Lemieux MJ, Song J, et al. Structure and mechanism of the glycerol-3-phosphate transporter from E. coli. Science. 2003;301:616-620.
Malandro MS, Kilberg MS. Molecular biology of mammalian amino acid transporters. Annu Rev Biochem. 1996;65:305-336.
Maloney PC. Bacterial transporters. Curr Opin Cell Biol. 1994;6:571-582.
Muakami S, Nakashima R, Yamashita E, Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419:587-593.
Nury H, Dahout-Gonzalez C, Trézéguet V, et al. Relations between structure and function of the mitochondrial ADP/ATP carrier. Annu Rev Biochem. 2006;75:713-741.
Orlowski J, Grinstein S. Na+/H+ exchangers of mammalian cells. J Biol Chem. 1997;272:22373-22376.
Pebay-Peyroula E, Dahout-Gonzalez C, Kahn R, et al. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatrachtyloside. Nature. 2003;426:39-44.
Walmsley AR, Barrett MP, Bringaud F, Gould GW. Sugar transporters from bacteria, parasites and mammals: Structure-activity relationships. Trends Biochem Sci. 1998;23:476-480.
Wright EM, Loo DDF, Turk E, Hirayama BA. Sodium cotransporters. Curr Opin Cell Biol. 1996;8:468-473.
Yernool D, Boudker O, Jin Y, Gouaux E. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature. 2004;431:811-818.
Yu EW, McDermott G, Zgurskaya HI, et al. Structural basis of multiple drug-binding capicity of the AcrB multidrug efflux pump. Science. 2003;300:976-980.