CHAPTER 8 Membrane Pumps
Membrane Permeability: An Introduction
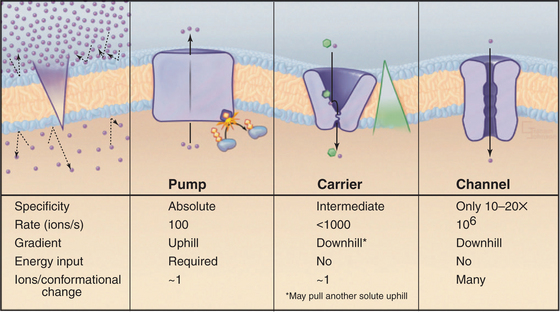
Although lipid bilayers provide a barrier to diffusion of ions and polar molecules larger than about 150 D, protein pores provide selective passages for ions, and other larger molecules across membranes. Integral proteins that control membrane permeability fall into three broad classes—pumps, carriers, and channels—each with distinct properties (Fig. 8-1). These proteins allow cells to control solute traffic across membranes, an essential feature of many physiological processes.
This chapter and Chapters 9, 10, and 11 consider, in turn, the three classes of proteins that control membrane permeability. Pumps are discussed first because they create the solute gradients required for the function of carriers and channels. The concluding chapter in this section, Chapter 11, illustrates how pumps, carriers, and channels work together to perform a remarkable variety of functions. An impor-tant point is that differential expression of a subset of isoforms of these proteins in specific membranes allows differentiated cells to perform a wide range of complex functions.
Membrane Pumps
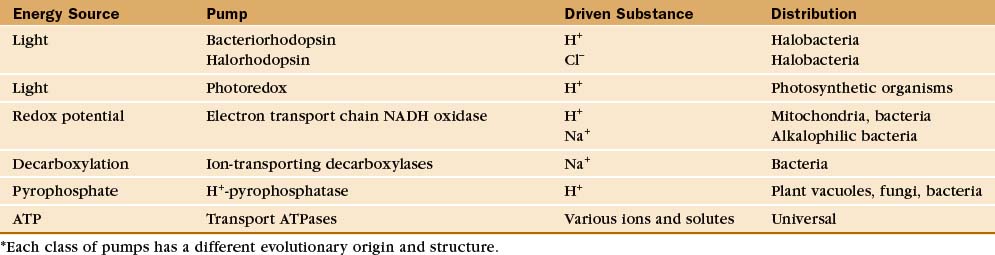
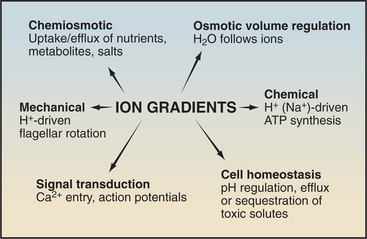
Protein pumps transport ions and other solutes across membranes up concentration gradients as great as 1 million-fold. Energy for this task can come from a variety of sources: light, oxidation-reduction reactions, or, most commonly, hydrolysis of ATP (Table 8-1). Energy is conserved in the form of transmembrane electrical or chemical gradients of the transported ion or solute. The potential energy in these ion gradients drives a variety of energy-requiring processes (Fig. 8-2). Most known biological pumps translocate cations. Although they could just as well move anions, cations were selected during the evolution of early life forms 3 billion years ago.
Pumps are also called primary active transporters because they transduce electromagnetic or chemical energy directly into transmembrane concentration gradients. Some carriers use ion gradients created by pumps to drive the uphill movement of other ions or solutes, so these are called secondary transporters (see Chapter 9). Channels are passive transporters, allowing net diffusion of ions and water only down their concentration gradients (see Chapter 10).
Diversity of Membrane Pumps
A vast array of integral membrane proteins can capture energy from an external source to pump ions and other solutes across biological membranes (Table 8-1). The protein families differ in their energy sources and transported materials. Fortunately, these pumps had a limited number of common ancestors, providing a relatively simple classification and generalizations about their structures and mechanisms. Given the importance of pumps in establishing transmembrane electrochemical gradients, the simplicity of this list is remarkable. Its brevity may be attributable to the fact that a single pump can drive a whole host of secondary reactions mediated by different carriers.
This chapter considers four representative pumps, emphasizing examples in which both high-resolution structures and detailed biochemical analysis of pathways are available. Chapter 19 provides additional details on H+ translocation by redox-driven cytochrome c oxidase and the role of F-type pumps in ATP synthesis by mitochondria and chloroplasts. Microbiology texts provide more information on pumps driven by decarboxylases and pyrophosphatases.
Light-Driven Proton Pumping by Bacteriorhodopsin
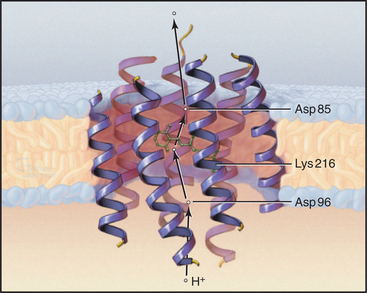
Owing to its simplicity, its small size, and the availability of a high-resolution structure (Fig. 8-3), more is known about light-driven transport of protons by bacteriorhodopsin than about any other pump. This pump allows the halophilic (salt-loving) Archaea Halobacterium halobium to convert light energy into a proton gradient across its plasma membrane. The 26-kD pump packs into two-dimensional crystalline arrays in the plasma membrane. The polypeptide is folded into seven α-helices that cross the lipid bilayer. The light-absorbing chromophore retinal (vitamin A aldehyde) is bound covalently to the side chain of lysine 216 (Lys216) via a Schiff base. This chromophore makes the protein and the membrane purple.
Bacteriorhodopsin absorbs light and uses the energy to pump protons out of the cell. A proton-driven ATP synthase uses this proton gradient to make ATP (Fig. 8-5). The proton pathway includes the side chains of aspartate 96 (Asp96), aspartate 85 (Asp85), glutamate 204 (Glu204), and the Schiff base. Local environments give the two aspartates remarkably different ionization constant (p Ka) values. Asp96 has a very high p Ka of about 10, so it can serve as a proton donor. Asp85 has a low p Ka of about 2, so it serves as a proton acceptor. Absorption of a photon changes the conformation of the retinal and the p Ka of the Schiff base. These four groups work together to transfer a single proton from the cytoplasm to the extracellular space.
The net result of this cycle is rapid vectorial transport of a proton from the cytoplasm out of the cell. Steps 4 to 6 are rate limiting, occurring at a rate of about 100 s−1. The other reactions are fast, provided that there is an adequate flux of light. Retinal not only captures energy by absorbing a photon but also acts as a switch that changes both the accessibility and affinities of the proton-binding groups in a sequential fashion.
In addition to bacteriorhodopsin, halobacterial plasma membranes contain two related proteins: halorhodopsin and sensory rhodopsin. Halorhodopsin absorbs light and pumps chloride into the cell. Interestingly, a single amino acid substitution can reverse the direction of pumping. Sensory rhodopsin couples light absorption by its bound retinal to phototaxis (swimming toward light) with a tightly coupled transducer protein. In the absence of this transducer, sensory rhodopsin transports protons out of the cell much like bacterial rhodopsin. The design of these seven-helix transporters is remarkably similar to that of the large family of seven-helix receptors, especially the photoreceptor proteins that vertebrates use for vision (see Fig. 24-2).
ATP-Driven Pumps
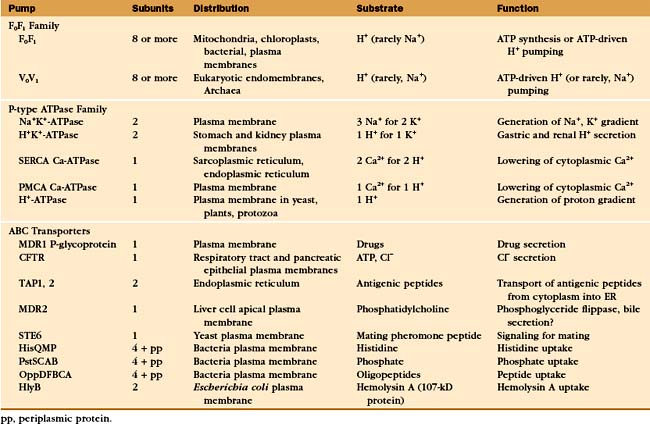
Three families of transport ATPases (Table 8-2) are essential for the physiology of all forms of life. F0F1-ATPases and P-type ATPases differ in structure, but both generate electrical and/or chemical gradients across membranes. ABC transporters not only produce ion gradients but also transport a much wider range of solutes across membranes. Chemical inhibitors have been useful in characterizing these pumps, and some are also used therapeutically (Table 8-3).
| Agent | Target |
|---|---|
| Cardiac glycosides* (e.g., ouabain, digitalis) | Na+K+ -ATPase |
| Omeprazole* | H+K+ -ATPase (parietal cell) |
| Oligomycin | F0F1-ATP synthase |
Free energy released by ATP hydrolysis puts a limit on the concentration gradient that these pumps can produce. If transport is electrically neutral (i.e., if it does not produce a membrane potential; see Fig. 10-17), the maximum gradient is about 1 million-fold. Such an extraordinary gradient is actually created by the P-type, electrically neutral H+K+−ATPase of gastric epithelial cells, which acidifies the stomach down to a pH of 1.
F0F1-ATPase Family
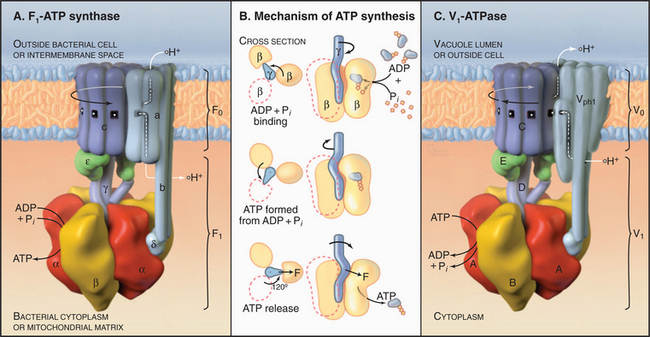
The two major subdivisions of this family are called F0F1-ATPases (or F-type ATPases) and V0V1-ATPases (or V-type ATPases) (Figs. 8-4 and 8-5). V0V1-ATPases, named for their location in the vacuolar system of eukaryotes, pump protons into organelles and out of Archaea. F0F1-ATPases of Bacteria, mitochondria, and chloroplasts generally run in the opposite direction, using proton gradients generated by other membrane proteins to drive ATP synthesis. However, purified F0F1-ATPases are freely reversible, using ATP hydrolysis to pump protons or alternatively proton gradients to synthesize ATP. Hence, these enzymes are called both ATP-synthases and F-type ATPases.
Phylogenetic analysis of the subunit polypeptides traces the origin of V-type ATPases to the precursor of all contemporary life forms (see Fig. 1-1). The genes for F-type ATPase subunits arose by divergence in Bacteria after they separated from Archaea. Eukaryotes came to have both F-type ATPases and V-type ATPases when symbiotic Bacteria gave rise to mitochondria and chloroplasts. Two subtle points are of interest here. First, a few Bacteria still have a V-type ATPase. Second, in contrast to the situation in eukaryotes, archaeal V-type ATPases function as ATP synthases similar to mitochondrial and bacterial F-type ATP synthases.
F-type ATPases (ATP Synthases)
F-type ATPases of mitochondria, chloroplasts, and bacterial plasma membranes produce most of the world’s ATP during aerobic metabolism (see Chapter 19). Redox-driven and light-driven pumps create proton gradients to drive ATP synthesis by F-type ATPases. When required by circumstances, many Bacteria use their F-type ATPase to produce a proton gradient at the expense of ATP hydrolysis. Eukaryotes have elaborate mechanisms to inactivate the ATPase and prevent futile cycles of ATP synthesis and hydrolysis. For example, in mitochondria, an inhibitory protein binds the F1-ATPase if the oxygen supply required to generate the proton gradient is compromised.
F0F1-ATPase has two parts (Fig. 8-5). Water-soluble, globular F1 catalyzes ATP hydrolysis or synthesis. F0 is embedded in the membrane and passively conducts protons across the lipid bilayer. A stalk connects F1 to F0, providing a way to couple proton translocation to ATP synthesis. Given a higher concentration of protons outside a Bacterium or mitochondrion than inside, protons pass through F0 and drive the synthesis of ATP by F1. Conversely, in bacteria, ATP hydrolysis by F1 can drive protons out of the cell.
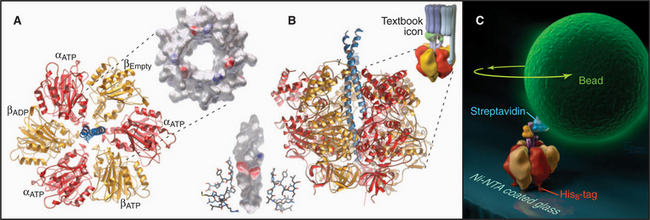
Pioneering biochemical studies and the crystal structure of F1 (Fig. 8-4) suggested that rotation of a protein shaft couples proton fluxes in F0 to ATP synthesis or, alternatively, couples ATP hydrolysis in F1 to proton pumping by F0. In the simplest case, bacterial F1 consists of five different types of polypeptides in the ratio a3b3gDe. Mitochondrial F1 has additional subunits. The α- and β-subunits are folded similarly and arranged alternately like segments of a orange. The γ-subunit is folded into a long, antiparallel, α-helical coiled-coil that forms a shaft. This hydrophobic shaft fits tightly in a hydrophobic sleeve in the middle of the hexamer of α- and β-subunits. To accommodate the asymmetrical shaft, each of the surrounding α- and β-subunits has a slightly different conformation. Each α- and β-subunit has an adenine nucleotide-binding site at the interface with its neighbor. ATP bound stably to α-subunits does not participate in catalysis. Nucleotide-binding sites on β-subunits catalyze ATP synthesis and hydrolysis.
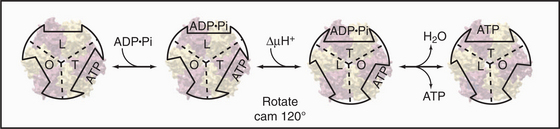
Mechanical rotation of the γ-subunit inside F1 is tightly coupled to ATP synthesis or hydrolysis. A proton gradient across the membrane can drive a flux of protons through the F0 complex. This drives clockwise (when viewed from F0) rotation of the γ-subunit inside the ab hexamer, like the camshaft in a motor. The mechanical force produced by the asymmetric camshaft drives conformational changes in β-subunits that synthesize ATP (Figs. 8-5 and 8-6). When the machine operates in the other direction, ATP hydrolysis drives counterclockwise rotation of the shaft, which can pump protons through F0.
Light microscopy is used to observe rotation directly (Fig. 8-4C). F1 is attached to a glass coverslip, and a tiny bead or actin filament is attached to the free end of the γ-subunit. ATP hydrolysis by the β-subunits drives the rotation of the bead or filament on the γ-subunit. If the bead is magnetic, a rotating magnet field can be used to drive the shaft and synthesize ATP from ADP and phosphate. The γ-subunit rotates at a maximum rate of about 130 times per second (8000 rpm) in mitochondria and twice as fast in chloroplasts.
V-Type ATPases
Vacuolar ATPases (Fig. 8-5) are found in the membranes bounding acidic compartments in eukaryotic cells, including clathrin-coated vesicles, endosomes, lysosomes (see Chapter 22), Golgi apparatus (see Chapter 21), secretory vesicles (including synaptic vesicles), and plant vacuoles. V-type pumps are also present in the plasma membranes of cells specialized to secrete protons, such as osteoclasts (see Fig. 32-6), macrophages, and kidney tubule intercalated cells.
V-type pumps have two functions. First, they acidify all the compartments listed here using rotation of the c-subunits to drive proton translocation. The acidic pH promotes ligand dissociation from receptors in endosomes and activates lysosomal hydrolases, as well as many other reactions (see Chapter 22). Second, proton gradients across these membranes provide the energy source to drive H+−coupled transport of other solutes by carriers, such as the uptake of neurotransmitters by synaptic vesicles (see Figs. 11-8 and 11-9).
P-Type Cation Pumps: e1E2-ATPases
All living organisms depend on P-type ATPases (Table 8-2) to pump cations across membranes. Their name comes from the fact that they utilize a high-energy covalent β-aspartyl phosphate intermediate. They are also called e1E2-ATPases from a description of the conformational changes that they undergo during the course of their mechanism.
Eukaryotic P-type ATPases generate primary ion gradients across the plasma membrane that are required for the function of ion channels (see Chapter 10) and most cation-coupled transport mediated by carrier proteins (see Chapter 9). In animal cells, Na+K+-ATPase produces the primary gradients of Na+ and k+. In plants and fungi, the functional homolog H+−ATPase generates a proton gradient. Production of these primary ion gradients is expensive, consuming up to 25% of total cellular ATP. Other eukaryotic P-type ATPases acidify the stomach and clear cytoplasm of the second messenger, Ca2+ (see Fig. 26-12). Bacterial P-type ATPases scavenge k+ and Mg2+ from the medium and export Ca2+, Cu2+, and toxic heavy metals.
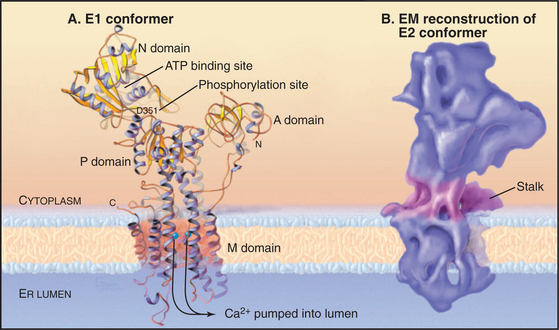
The P-type ATPase that is best understood is the sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA1), which pumps Ca2+ out of the cytoplasm into the endoplasmic reticulum (Fig. 8-7). ATP hydrolysis provides energy to move Ca2+ across the membrane up a steep concentration gradient. The mechanism is understood in detail, thanks to extensive biochemical analysis and crystal structures of five of the chemical intermediates along the pathway (Fig. 8-8). This analysis was possible because the enzyme is abundant in the sarcoplasmic reticulum of skeletal muscle (see Fig. 39-10), allowing it to be purified in large quantities.
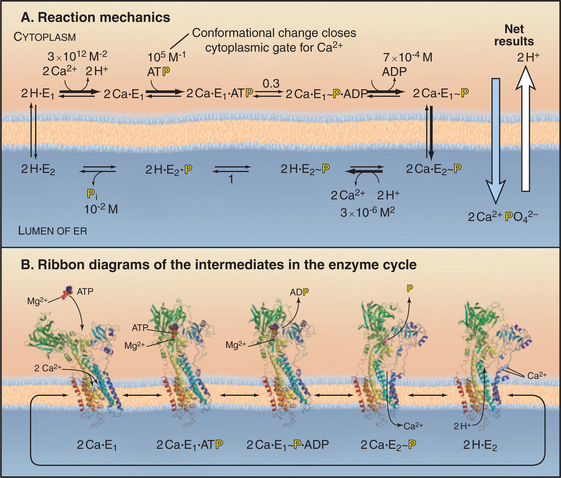
Figure 8-8 Reaction mechanism of the sarcoendoplasmic reticulum calcium–atpase. A, Biochemical pathway. The E stands for enzyme, having two conformations: e1 and e2. e1 has the Ca2+−binding site oriented toward the cytoplasm. e2 has the Ca2+−binding site oriented toward the lumen of the endoplasmic reticulum. Ca2+ binds e1 on the cytoplasmic side. Subsequent binding of ATP and phosphorylation of the enzyme drive the enzyme toward the e2 state and transport Ca2+ up a steep concentration gradient into the lumen of the endoplasmic reticulum. Dephosphorylation of the enzyme favors a return to the e1 state.B, Ribbon diagrams of structures along the biochemical pathway. Conformational changes are coupled to ATP hydrolysis and transport of Ca2+. Starting on the left side of part A, the pump without bound Ca2+ or ATP vacillates between the e2 and e1 conformations, alternatively exposing the Ca2+−binding sites on the two sides of the membrane. If Ca2+ ions are available in the cytoplasm, such as after activation of muscle (see Fig. 39-16), they bind cooperatively to the e1 conformation with micromolar affinity. Ca2+ binding strongly stimulates enzyme activity by favoring Mg-ATP binding to the N-domain. Mg-ATP can bind simultaneously to both the N- and the P-domains, so its presence favors a striking change in conformation that requires the N-domain to rotate and the A-domain to pull on a transmembrane helix that closes a gate between the bound Ca2+ and the cytoplasm. This conformation brings the γ-phosphate of ATP into proximity with the side chain of Asp351 and allows formation of the phosphoenzyme intermediate. The equilibrium constant for phosphorylation is near unity, so most of the energy from ATP hydrolysis is stored in a high-energy conformation of the protein. After transfer of γ-phosphate to the enzyme, ADP dissociates. This results in a rotation of the A-domain that moves several transmembrane helices to expose the Ca2+−binding sites to the lumen of the endoplasmic reticulum and reduces the affinity for Ca2+ by several orders of magnitude. Thus Ca2+ dissociates into the lumen, completing its uphill transfer from the cytoplasm. Hydrolysis of the phosphorylated intermediate and dissociation of phosphate reverse the conformational changes in both the cytoplasmic and transmembrane domains, completing the cycle.
(References: Toyoshima C, Nomura H, Tsuda T: Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues. Nature 432:361–368, 2004; and Soerensen T, Moeller JV, Nissen P: Phosphoryl transfer and calcium ion occlusion in the calcium pump. Science 304:1672–1675, 2004. PDB files: 1EUL, 1T5S, 1T5T, IWPG, IIWO.)
The 100-kD Ca2+−ATPase consists of two regions. Ten α-helices cross the membrane bilayer and bind two Ca2+ ions side by side in the middle of the membrane. The globular region in the cytoplasm consists of three domains. The N-domain binds ATP and transfers its γ-phosphate to aspartic acid 351 (Asp351) in the P-domain. The A-domain transmits large conformational changes in the cytoplasmic domains to the transmembrane helices, which (6 of 10 helices) alternate between two conformations. The e1 conformation allows access to the Ca2+ binding sites from the cytoplasm (Fig. 8-8A). The e2 conformation allows access from the lumen of the endoplasmic reticulum (Fig. 8-8B). Each step along the pathway—Ca2+ binding, ATP binding, phosphorylation of the enzyme, dissociation of ADP, and hydrolysis of the β-aspartyl phosphate intermediate—causes linked conformational changes in both the cytoplasmic domains and six of the transmembrane helices. The changes in the transmembrane domain alters the affinity of the protein for Ca2+ and the exposure of Ca-binding sites on the two sides of the membrane. Figure 8-8 describes the cycle of ATP hydrolysis and Ca2+ transport in detail. Note that the transition between the e1 and e2 conformations involves an “occluded” state in which the bound Ca2+ is not accessible on either side of the membrane. This occluded state allows the pump to transport against a large concentration gradient without a leak.
Because the cycle transfers two Ca2+ into the lumen and two H+ out, it generates an electrical potential (see Chapter 10). In the steady state, the Ca2+ gradient is maintained, but the H+ gradient dissipates, owing to H+ permeability across the membrane and to the buffering capacity of the lumen. All reactions in the pathway are reversible, so a large gradient of Ca2+ across the membrane can drive the synthesis of ATP.
All P-type ATPases consist of homologous α-subunits with large cytoplasmic domains and a minimum of six transmembrane helices. Eukaryotic P-type ATPases, such as the Na+K+− and Ca2+−ATPase s have 10 transmembrane helices. Many P-type ATPase are about 110 kD, like the Ca-ATPase, but some are larger or smaller, owing to variable features. Na+K+−ATPase and H+K+−ATPase require a 50-kD glycosylated β-subunit with a single transmembrane segment for both transport and intracellular trafficking.
Some P-type ATPases are involved in human diseases. Mutations in Ca2+−ATPase cause muscle stiffness and cramps. Mutations in Cu2+−ATPase s cause two inherited diseases: Menkes’ syndrome, in which patients are copper-deficient owing to impaired intestinal absorption, and Wilson’s disease, in which the inability to remove copper from the liver is toxic. Omeprazole and related drugs are used to treat ulcers by inhibiting gastric H+K+−ATPase. Drugs called cardiac glycosides strengthen the heartbeat by inhibiting the cardiac isoform of Na+K+−ATPase (see Fig. 11-13 for details). These drugs, which derive originally from the foxglove plant, were among the first used to treat congestive heart failure.
ABC Transporters
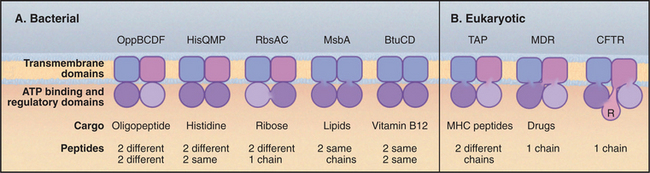
ABC transporters form the largest and most diverse family of ATP-powered pumps (Table 8-2). They are found in all known organisms, so the founding gene must have originated in the common ancestor of all living things. The genome of baker’s yeast encodes at least 30 ABC transporters, compared with 16 P-type ATPases, one F-type ATPase, and one V-type ATPase. ABC transporters are the largest gene family in the colon bacterium Escherichia coli. In eukaryotes particular family members are located in the plasma membrane, endoplasmic reticulum, and, most likely, other membranes.
ABC transporters have a modular design that includes two transmembrane domains and two cytoplasmic domains that hydrolyze ATP (Fig. 8-9A). Each transmembrane domain consists of a bundle of α-helices that spans the bilayer: typically six times but up to ten times in some examples. Two sequences in the nucleotide-binding domain give the family its name (ATP-binding cassette). The Walker A motif (GXXGXGKS/T, where X is any residue) is also called a P loop, since it binds the γ-phosphate of ATP in ABC transporters and other ATP-binding proteins. The Walker B motif (RX6–8F4D, where F is any hydrophobic residue) interacts with the Mg2+ bound to ATP. Motif B is typically separated from motif A by 100 to 150 residues along the sequence.
Various “experiments” of nature during evolution show that the four independently folded domains of an ABC transporter can be assembled by association of up to four subunits or by folding of a single polypeptide (Fig. 8-9). Gram-negative bacterial transporters often utilize periplasmic subunits that bind and concentrate transported substrates in the vicinity of the pump. Some vertebrate ABC transporters include an additional cytoplasmic R-domain in the single polypeptide for regulation by phosphorylation.
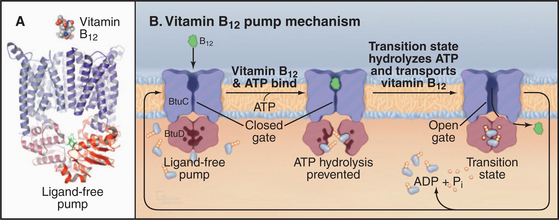
The crystal structure of the E. coli BtuCD vitamin B12 transporter (Fig. 8-10) suggests how ABC transporters might work. The molecule consists of four subunits: two copies of the transmembrane BtuC subunit and two copies of the nucleotide-binding BtuD subunit. Each transmembrane subunit consists of ten transmembrane helices. The interface between the BtuC subunits forms a large chamber open outside the cell (the periplasmic space in this case). The chamber is lined by hydrophobic side chains that are expected to interact with vitamin B12. The chamber penetrates more than halfway across the lipid bilayer but is blocked on the cytoplasmic side by residues that form a gate. The nucleotide-binding cytoplasmic subunits form large interfaces with their partner transmembrane subunits and have a small but highly conserved interface with the other nucleotide-binding subunit. This small interface positions ATP-binding sites of the two subunits next to each other. A periplasmic protein binds vitamin B12 and presents it to the transporter.
Other family members are more problematic. The vertebrate cystic fibrosis transmembrane conductance regulator (CFTR [Fig. 8-9B]) looks like a pump but acts like a channel. It allows Cl− to move down its concentration gradient out of the cell. ATP binding and hydrolysis by the nucleotide-binding domains may open and shut this channel. Mutations in CFTR are responsible for cystic fibrosis (see Fig. 11-4). Of more than 1000 CFTR mutations known to cause cystic fibrosis, by far the most common mutation is deletion of phenylalanine 508. This position corresponds to a highly conserved hydrophobic residue in the vitamin B12 transporter that is important for the interaction of BtuD with BtuC. Mutant DF508 CFTR misfolds and is retained in the ER and destroyed, depriving the plasma membrane of chloride channel activity. Depleting Ca2+ from the ER by inhibiting the SERCA Ca2+ ATPase can apparently allow DF508 CFTR to escape from calcium-dependent chaperones (see Fig. 20-10) and function on the cell surface.
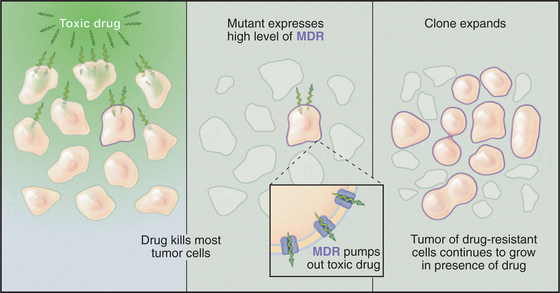
The multiple drug resistance proteins (MDR1 and MDR2) are ABC transporters that provide a challenge for cancer chemotherapy (Fig. 8-11). In about half of the cases in which chemotherapy fails to cure cancer in humans, the cause is the emergence of clones of tumor cells that overexpress an MDR. Normal cells use a low level of MDR1 to export unknown substrates, perhaps a steroid, a phospholipid, or another hydrophobic molecule. MDR can also transport many hydrophobic compounds, including some chemotherapeutic drugs. These drugs enter cells by dissolving in the membrane, and they subsequently poison vital cellular processes. Cells that overexpress MDR survive by pumping the drug out of the cell.
Some even less conventional ABC transporters appear to regulate ion channels. The sulfonylurea receptor (SUR) is an ABC transporter required for the function of an ATP-sensitive potassium channel (KATP) that regulates insulin secretion. SUR binds drugs called sulfonylureas that are used to treat forms of diabetes involving inadequate insulin secretion. These drugs activate secretion by inhibiting the kATP channel (see Chapter 10).
Abrahams JP, Leslie AGW, Lutter R, Walker JE. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621-628.
Borst P, Oude Elferink R. Mammalian ABC transporters in health and disease. Annu Rev Biochem. 2002;71:537-592.
Cross RL. Turning the ATP motor. Nature. 2004;427:407-408.
Davidson A. Not just another ABC transporter. Science. 2002;296:1038-1040.
Davidson AL, Chen J. ATP-binding cassette transporters in bacteria. Annu Rev Biochem. 2004;73:241-268.
Facciotti MT, Rouhani-Manshadi S, Glaeser RM. Energy transduction in transmembrane ion pumps. Trends Biochem Sci. 2004;29:445-451.
Kaplan JH. Biochemistry of the Na, K-ATPase. Annu Rev Biochem. 2002;71:511-535.
Kinosita KJr, Adachi K, Itoh H. Rotation of F1-ATPase. How an ATP-driven molecular machine may work. Annu Rev Biophys Biomol Struct. 2004;32:245-268.
Kuhlbrandt W. Biology, structure and mechanism of P-type ATPases. Nat Rev Mol Cell Biol. 2004;5:282-295.
Lancaster CRD. Ion pumps in the movies. Nature. 2004;432:286-287.
Lanyi JK. Bacteriorhodopsin. Annu Rev Physiol. 2004;66:665-688.
Locher KP. Structure and mechanism of ABC transporters. Curr Opin Struct Biol. 2004;14:426-431.
Locher K, Lee A, Rees D. The E. coli BtuCD structure: A framework for ABC transporter architecture and mechanism. Science. 2002;296:1091-1098.
Nishizaka T, Oiwa K, Noji H, et al. Chemomechanical coupling in F1-ATPase revealed by simultaneous observation of nucleotide kinetics and rotation. Nat Struct Mol Biol. 2004;11:142-148.
Oster G, Wang H. Rotary protein motors. Trends Cell Biol. 2003;13:114-121.
Soerensen T, Moeller JV, Nissen P. Phosphoryl transfer and calcium ion occlusion in the calcium pump. Science. 2004;304:1672-1675.
Subramaniam S, Hirai T, Henderson R. From structure to mechanism: Electron crystallographic studies of bacteriorhodopsin. Phil Trans A Math Phys Eng Sci. 2002;360:859-874.
Toyoshima C, Inesi G. Structural basis of ion pumping by Ca2+-ATPase of the sarcoplasmic reticulum. Annu Rev Biochem. 2004;73:269-292.
Toyoshima C, Nomura H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature. 2002;418:605-611.
Toyoshima C, Nomura H, Tsuda T. Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues. Nature. 2004;432:361-368. (The journal web site associated with this paper has a movie of the structural changes during the ATPase cycle: http://www.nature.com/nature/journal/v432/n7015/suppinfo/nature02981.html.).