CHAPTER 45 Meiosis*
Meiosis (from the Greek, meaning “reduction”) is a specialized program of two coupled cell divisions used by eukaryotes to maintain the proper chromosome number for the species during sexual reproduction. The number of chromosomes is halved in meiosis; therefore, the subsequent fusion of male and female gametes restores the proper chromosome number for the species. The reduction in chromosome number is achieved by randomly separating homologous chromosomes, each pair of which is composed of one chromosome donated by the mother and one donated by the father. This pairing and subsequent separation of homologous chromosomes are typically made possible by genetic recombination, which occurs during the lengthy and complex prophase of the first meiotic division. The random segregation of homologous chromosomes and the genetic recombination that make this possible form the physical basis of the laws of classical genetics, first proposed by Gregor Mendel in 1866.
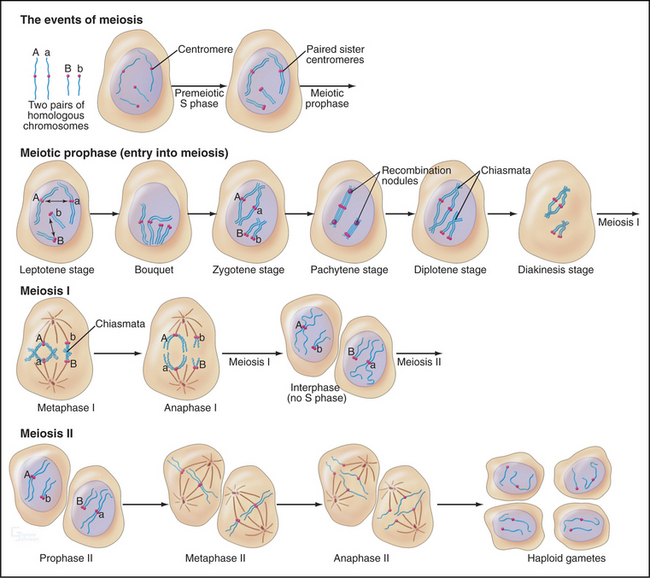
The unique events of meiosis occur in the first division, termed meiosis I (Figs. 45-1 and 45-2). Because the daughter cells have half the number of chromosomes, meiosis I is also known as the reductional division. The second division, meiosis II, is similar in most respects to mitosis: Sister chromatids segregate from each other, and the number of chromosomes remains the same (Box 45-1; see also Chapter 44). Meiosis II is called the equational division. Meiosis is an ancient process that occurs in virtually all higher eukaryotes, including the animal, fungal, and plant kingdoms.
BOX 45-1 Important Differences between Meiosis and Mitosis
The products of meiosis are haploid. The products of mitosis are diploid.
Meiosis: An Essential Process for Sexual Reproduction
Without meiosis, there would be no sex because every fusion of gametes would increase the number of chromosomes in the progeny. Sexual reproduction is an important survival strategy that offers organisms a mechanism for altering the genetic makeup of offspring. This strategy has been conserved throughout higher eukaryotes and is inextricably linked with the mechanism of meiosis.
Meiosis I also produces novel versions of chromosomes by exchange of DNA segments between homologs. This occurs because each chromosome must typically undergo at least one genetic recombination (crossover) event to segregate properly at anaphase of meiosis I. If the chromosomes of all the individuals of a species were identical, meiosis and sexual reproduction would only provide different combinations of the same chromosomes. However, human chromosomes vary between individuals, averaging about one difference (polymorphism) per 1000 base pairs, and it is estimated that, overall, at least 106 sites across the genome have variant versions. Recombination involves exchange of chromosomal segments, producing new chromosomes that are a patchwork of segments from the maternal and paternal homologs. The combined effects of recombination and random assortment of homologs in meiosis I yields a vast number of different gametes and provides an important source of genetic diversity that permits eukaryotic populations to adapt to changing environmental conditions.
The Language of Meiosis
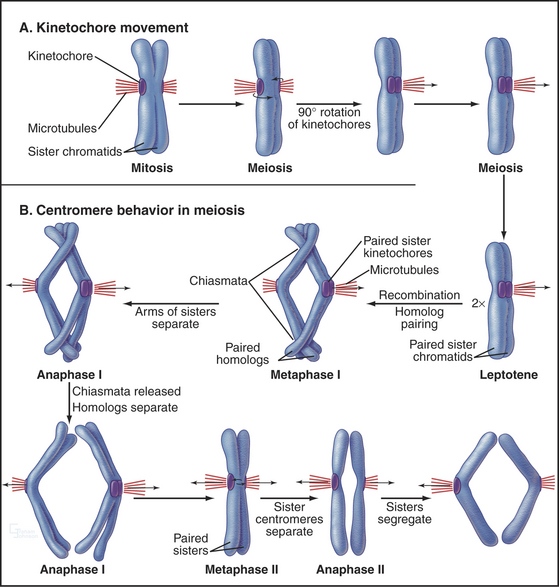
Meiosis is all about the segregation of the paired homologous chromosomes. This process shows some key differences from mitosis (Box 45-1). When the homologs are balanced at the metaphase plate of the meiosis I spindle, it is the chiasmata that hold them together and counteract the pulling force of the spindle on the kinetochores (Fig. 45-2). Cohesion between the chromatid arms holds chiasmata in place until it is released at anaphase of meiosis I. Centromeres of the sister chromatids remain associated with one another throughout meiosis I until anaphase of meiosis II. This means that at anaphase, when the chiasmata are released, each pair of sister chromatids migrates to the same spindle pole. As a result, the progeny of meiosis I have the haploid number of chromosomes each paired with a sister chromatid. Box 45-2 reviews some genetics terms that are helpful in understanding meiosis.
BOX 45-2 Brief Overview of Genetic Terminology
A comprehensive introduction to the field of genetics is beyond the scope of this text. However, here are a number of terms used by geneticists that will assist in the understanding of the discussion of genetic recombination and its role in meiosis (also see Box 6-2).
Two types of recombination events occur during meiosis (Fig. 45-3). The first of these—noncrossover events (frequently referred to as gene conversion)—may involve the loss of one or more genetic markers. Noncrossover events are the most common outcome of the programmed double-strand DNA breaks that occur during leptotene. They are thought to involve the invasion of a double helix by a region of single-stranded DNA with complementary sequence but then ejection of this sequence before assembly of a Holliday junction and completion of recom-bination.
Recombination
Because recombination is the key to the behavior of chromosomes in meiosis I, this process is discussed in detail herein to provide a mechanistic underpinning for understanding later events. Meiotic recombination is very similar to the process of homologous recombinational repair of double-strand DNA breaks in somatic cells (review Box 43-1 and Fig. 43-15 as a prelude to studying meiotic recombination).
Spo11 generates programmed double-strand DNA breaks very early during meiotic prophase (Fig. 45-3). Spo11 is a type II DNA topoisomerase (see Chapter 13, under the section titled “Proteins of the Mitotic Chromosome and Chromosome Scaffold”) that cleaves both DNA strands in a reaction that produces a covalent linkage between a tyrosine on the enzyme and the cleaved phosphodiester backbone. Where it has been measured, Spo11 creates about threefold to fivefold more DNA breaks than ultimately complete the recombination pathway to produce reciprocal exchanges of DNA between homologous chromosomes, or crossovers. An alternative pathway is thought to process the excess breaks, producing noncrossover events (Box 45-2 and Fig. 45-3I-J). Each pair of homologous chromosomes thus undergoes many noncrossover events and a very few crossover events (often only one) during meiosis I prophase.
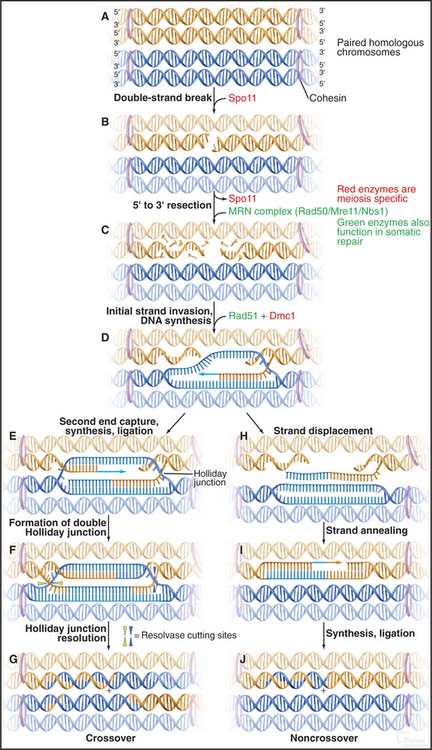
Figure 45-3 the events of recombination. Recombination occurs between homologs rather than sisters. A, Paired homologous chromosomes. Sister chromatids are held tightly together by cohesin, shown here schematically as hoops. B, Spo11 makes a double-strand break. C, Resection of the break. D, First strand invasion. At this point, the pathway splits in two, one outcome leading to a crossover and the other to a noncrossover. Crossover pathway: E, The second resected strand invades its homologous partner. New DNA synthesis fills the gaps. F, The resulting molecule contains a double Holliday junction (see Fig. 43-15B). If the resolvase (nuclease) cuts the double Holliday junction asymmetrically as shown (i.e., one vertical and one horizontal cut), the result is a crossover (G). If the cuts are symmetrical, a noncrossover molecule is produced. Noncrossover pathway: H, In most cases, the invading DNA strand is ejected prior to stabilization and formation of a double Holliday junction. I, DNA gap-filling and ligation yield a noncrossover chromosome (J).
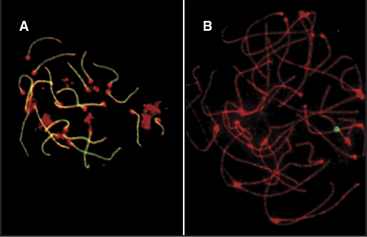
In both mice and yeast, double-strand breaks generated by Spo11 are required for normal segregation of homologous chromosomes. In Spo11-null mice, recombination is not initiated, and synapsis, if it occurs at all, is aberrant, often involving nonhomologous chromosomes (Fig. 45-4). In these mutant mice, spermatocytes die by apoptosis early in meiotic prophase, and oocytes die somewhat later. In contrast, the nematode Caenorhabditis elegans and the fruit fly Drosophila melanogaster do not require Spo11-induced double-strand breaks for synapsis of homologous chromosomes.
Once the DNA double-strand breaks have been produced, they are processed by the 5′ → 3′ exonuclease MRN (Mre11/Rad50/Nbs1), which chews back one strand of the double helix (a process called resection), leaving single-stranded tails at the 3′ end of the DNA molecules (Fig. 45-3C; see also Fig. 43-15). The same exonuclease functions in somatic DNA repair and in meiotic recombination.
Next, the single-stranded tails “invade” the other chromosomes, looking for complementary DNA se-quences. This process is driven by Rad51 and Dmc1, two proteins that are related to the E. coli RecA protein, which is essential for DNA recombination in bacteria. These proteins polymerize into nucleoprotein filaments on DNA and use ATP hydrolysis to catalyze homologous pairing and strand exchange reactions. The process inserts a single-stranded region of DNA into a double helix, displacing one of the two paired strands. Dmc1 functions only in meiosis, but Rad51 has other essential functions as well. Dmc1 may promote the search for homologous chromosomes, rather than sister chromatids as occurs in somatic DNA repair. Mutants that lack Dmc1 are defective in homologous chromosome pairing. Rad51p and Dmc1p are found in structures called early recombination nodules that are distributed along the chromosome axes early in meiosis (Fig. 45-9).
It is now believed that if only one single strand successfully invades the homologous chromosome, the outcome is a noncrossover event, whereas invasion of both single-strand tails leads to crossovers. The double invasion produces branched intermediates known as double Holliday junctions (Fig. 45-3F-G; see also Fig. 43-15B). These are then cleaved by as yet unknown nucleases and converted to mature crossover recombination products.
Tracking the Homologous Chromosomes through the Stages of Meiotic Prophase I
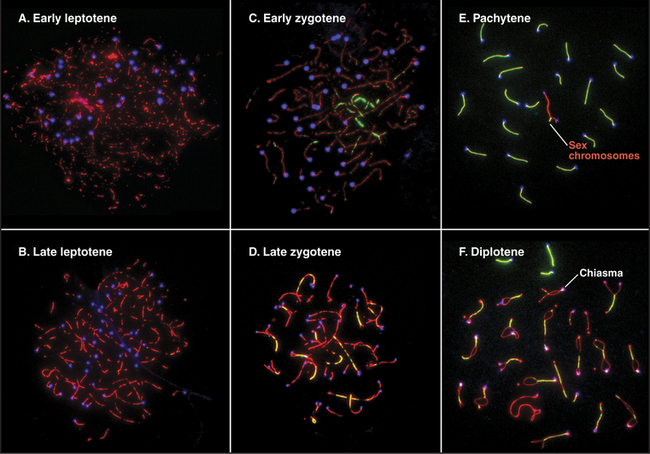
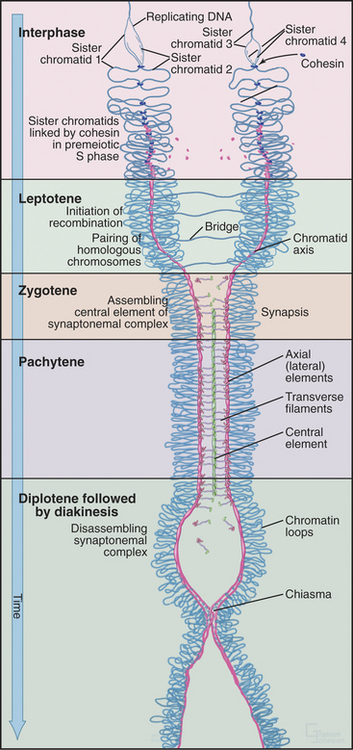
Pairing and recombination of homologous chromosomes take place during prophase of meiosis I. In the discussion of these processes, it is necessary to refer to the five stages of meiotic prophase: leptotene, zygotene, pachytene, diplotene, and diakinesis (Fig. 45-1). As the understanding of meiotic prophase advances, the significance of these stages is being reassessed. In particular, these morphologic stages do not correspond directly to the steps of meiotic recombination, as was assumed previously.
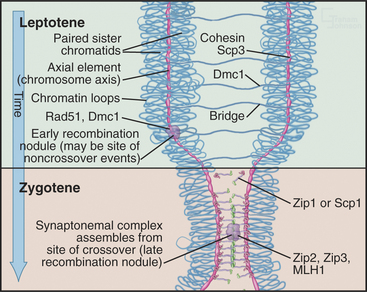
The start of leptotene (from the Greek, meaning “thin ribbon”) is defined by the first visible condensation of the chromosomes. Paired sister chromatids begin to condense as arrays of loops flanking a single dense protein-containing axis (Fig. 45-5A-B). This axis consists of proteins that play a role in mitotic chromosome structure as well as proteins that are specialized for meiotic chromosomes. For example, the cohesin com-plex is a prominent component of this axial struc-ture (see Fig. 13-19), but several of its components are replaced by meiosis-specific forms. According to recent models, recombination begins during leptotene with the formation of double-strand breaks, which are processed and a few selected for crossovers. By the end of leptotene, homologous chromosomes are aligned loosely about 400 nm apart (Fig. 45-6D-G).
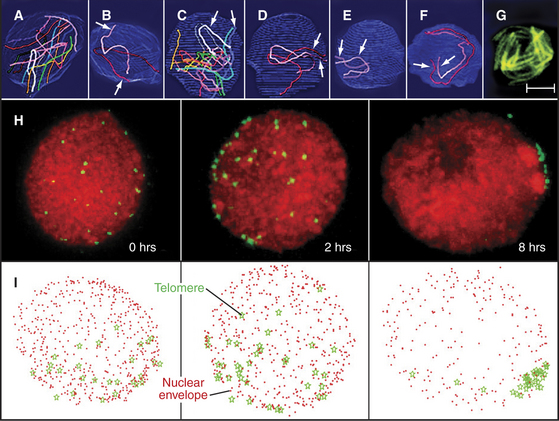
Figure 45-6 chromosomal movements during early meiotic prophase. a–g, pairing of homologous chromosomes during leptotene in the ascomycete sordaria. Scale bar is 1mm in A–F and 5mm in G. A–B, In early leptotene, homologous chromosomes (visualized in panels A–F by electron microscope reconstructions of serial-sectioned nuclei) are not yet aligned with one another. C–E, In mid-leptotene, regions of some homologs begin to align. (In panel D, only the telomeres have aligned. In panel E, the pair of homologs is fully aligned.) F, The alignment of homologs is complete by late leptotene. G, The alignment of homologs also can be seen by light microscopy using Spo76-GFP, a component of the chromosome axes. H–I, Stages of formation of the bouquet arrangement in rye. H, Telomeres (green) were detected in nuclei by in situ hybridization (see Fig. 13-15) after 0, 2, and 8 hours in culture. Chromatin is red. I, Three-dimensional models of the nuclei (nuclear periphery [red dots], telomere position [green stars]).
(A–G, Adapted from Tesse S, Aurora Storlazzi A, Kleckner N, et al: Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc Natl Acad Sci U S A 100:12865–12870, 2003. Copyright 2003 National Academy of Sciences, USA. H–I, Adapted from Carlton PM, Cowan CR, Cande WZ: Directed motion of telomeres in the formation of the meiotic bouquet revealed by time course and simulation analysis. Mol Biol Cell 14:2832–2843, 2003. Adapted from Molecular Biology of the Cell [Mol Biol Cell 14:2832–2843, 2003; published on-line before print as 10.1091/mbc. E02-11-0760] with the permission of The American Society for Cell Biology.)
During zygotene (from the Greek, meaning “yoke ribbon”), the next portion of prophase, homolog pairing, goes to completion in a process known as synapsis (Fig. 45-5C-D). This involves the assembly of a protein scaffold, the synaptonemal complex. Also in early zygotene, the telomeres cluster in a region of the nuclear envelope, giving rise to the “bouquet” arrangement of chromosomes (see next section). In pachytene (from the Greek, meaning “thick ribbon”), synapsis is complete, with the homologs joined together along their lengths by synaptonemal complex (Fig. 45-5E). During pachytene, crossovers are believed to mature into structures called chiasmata that will hold homologous chromosomes together through meiosis I metaphase.
Early in diplotene (from the Greek, meaning “double ribbon”), the synaptonemal complex disassembles, and chromosomes decondense (Fig. 45-5F). Later on, they start condensing again. Sister chromatids remain closely associated, whereas homologous chromosomes tend to separate from each other, held by the chiasmata. This part of meiotic prophase may last for days or years, depending on the sex and organism (up to 45 years or more in female humans).
In females, the chromosomes are very actively transcribed during diplotene, as the egg busily stores up materials for use during the first few divisions of embryonic development. In animal diplotene cells, the chromosomes have prominent loops and are known as lampbrush chromosomes (see Fig. 13-13). These loops are visible because the DNA is massively coated with nascent RNA transcripts and their associated proteins.
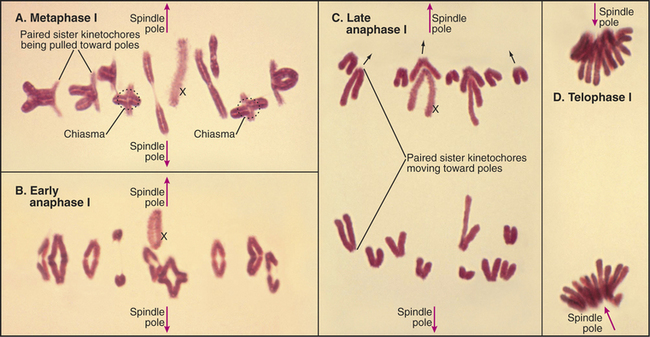
Diakinesis (from the Greek, meaning “across movement”) is the prometaphase of meiosis I. Following nuclear envelope breakdown, homologous chromosomes become particularly short and condensed. At metaphase I, the bivalents (pairs of homologous chromosomes) are aligned at a metaphase plate (Figs. 45-1 and 45-2). Each homolog (a pair of tightly linked sister chromatids) is attached to a single pole of the meiotic spindle. Chiasmata resist the pulling forces within the spindle. At anaphase I, the release of cohesion along the chromosome arms (but not between centromeres of sister chromatids) allows chiasmata to be resolved and homologs to move to opposite spindle poles. After telophase I, there is no DNA replication, and cells enter directly in the second meiotic division, which is mechanistically similar to mitosis. In the eggs of most female vertebrates, meiosis is arrested at metaphase II until fertilization.
Chromosomal Ikebana: The Bouquet Stage
During leptotene, the chromosomal telomeres attach apparently randomly to the surface of the nuclear envelope. As leptotene progresses, in most organisms these telomeres gradually move together to occupy the region of the nuclear envelope closest to the centrosome (spindle pole body in yeasts [Fig. 45-6]). In this region, chromosome axes attach to a dense plaque on the inner surface of the nuclear envelope. This clustering of telomeres in the nucleus requires the presence of microtubules in the cytoplasm. How this process is coordinated across the nuclear envelope is not known. Telomere clustering appears to be maximal at the leptotene-zygotene transition, when the chromosomes radiating into the nuclear interior resemble a bouquet of flowers, hence the name “bouquet stage.” The bouquet is a nearly universal feature of this phase of meiosis, although ironically, the popular model organisms C. elegans and D. melanogaster are exceptions.
Pairing and Synapsis in More Detail
Pairing describes the side-by-side alignment of homologous chromosomes at a distance. Homologs are paired in nonmeiotic cells in some organisms, such as the fruit fly D. melanogaster and budding yeast Saccharomyces cerevisiae, but not in vertebrates. As was mentioned earlier, pairing involves a search of homologous se-quences for one another. The entire genome is scanned during this process, because even when a single gene is transposed onto a different chromosome, this gene can still find its homologous partner. In certain organisms, such as C. elegans, pairing can involve specialized DNA sequences; however, this does not generally appear to be the case.
The earliest pairing events involve a tendency of homologous chromosome territories to move together in the nucleus even before leptotene chromosome condensation. The mechanism is unknown. Next, programmed double-strand breaks created by Spo11 initiate the recombination pathway during leptotene. In parallel, the condensing homologous chromosomes align with one another at a distance of about 400nm (Figs. 45-6 and 45-7). Genetic analysis in budding yeast revealed that mutants defective in the earliest stages of recombination are also defective in homolog pairing.
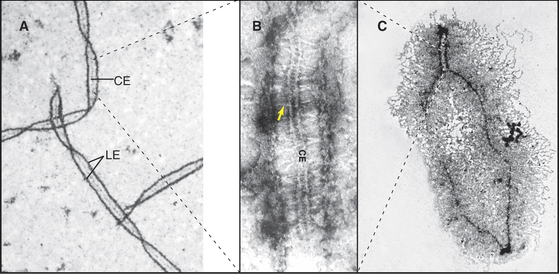
Homolog pairing initiated during leptotene becomes much more intimate during synapsis as the chromosomes become linked by transverse fibers to form the synaptonemal complex. This structure looks roughly like railroad tracks with a third rail running down the center (Figs. 45-7 and 45-8). The two outer rails, 90 to 100 nm apart, traditionally termed lateral elements, are the axes of the paired sister chromatids. For the sake of simplicity, this chapter refers to them as axial elements throughout meiosis I. Thin transverse filaments lying perpendicular to the axial elements appear to connect them to each other and to the central element (the “third rail”). Synaptonemal complex formation is initiated at a limited number of sites along the paired homologous chromosomes. These often correspond to sites where recombination events will mature into crossovers. Synapsis begins during zygotene, and by pachytene, a continuous synaptonemal complex is observed between homologous chromosomes (Fig. 45-5C-E).
It used to be thought that the synaptonemal com-plex aligns homologous chromosomes in preparation for recombination, but it is now clear that homolog pairing and the initiation of recombination precede synapsis. Furthermore, yeast mutants that affect synaptonemal complex formation do not affect pairing (i.e., homologous chromosomes pair but do not synapse), and under certain artificial circumstances, it is pos-sible for nonhomologous chromosomes to undergo synapsis.
Synaptonemal Complex Components
Both genetic and biochemical approaches have identified components of the synaptonemal complex. Perhaps the best studied is the budding yeast protein Zip1p, which is found in mature synaptonemal complex, between the axial elements (Fig. 45-9). Zip1p is predicted to have extensive regions of coiled-coil and is thought to assemble into a rod-shaped dimer. If the length of the Zip1p coiled-coil is altered, then synaptonemal complexes are produced in which the spacing between axial elements is altered. In zip1 mutants, recombination is initiated but fails to be completed at about 10% of sites. As a result, cells arrest late in prophase (see later discussion of the so-called pachytene checkpoint in the section titled “Cell-Cycle Regulation of Meiotic Events”). In mammals, a protein called Scp1 is localized in the transverse filaments. Scp1 has no sequence similarity to Zip1p, but both share a common organization: a coiled-coil flanked by two globular domains.
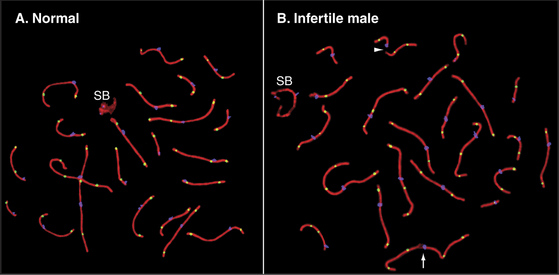
Several protein components of the axial elements (sister chromatid axes) have also been identified. One of these, Scp3, interacts with the cohesin complex (see later) and also with Rad51p and Dmc1p. In Scp3 knockout mice, the axial elements are much less prominent, and the axis of the condensed chromosome is about twofold longer. Other proteins of the synaptonemal complex, including Scp1, can still assemble, but chromosomes in male germ cells lack chiasmata and are unpaired. As a result, the germ cells die in pachytene/diplotene. It thus appears that in the mouse, Scp3 is required for axial condensation of meiotic chromosomes and for normal cohesion between sister chromatids, leading to formation of stable chiasmata. Human males who are mutant in Scp3 lack chiasmata, fail to segregate chromosomes normally in meiosis, and produce no viable sperm.
Chiasmata
The role of recombination in regulating chromosome dynamics during meiosis is most evident during the segregation of homologous chromosomes in meiosis I, mediated by chiasmata. Chiasmata (singular: chiasma) are specialized chromosomal structures that hold the homologous chromosomes together until anaphase I (Figs. 45-1 and 45-10). They are formed at sites where programmed DNA breaks generated by Spo11 undergo the full recombination pathway to generate crossovers.
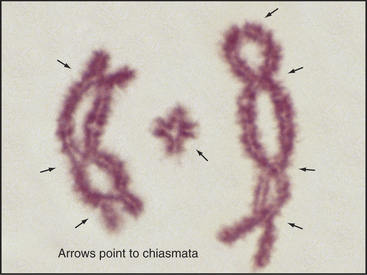
Figure 45-10 bivalents (paired homologous chromosomes) are held together by chiasmata after disassembly of the synaptonemal complex. Here, three diplotene bivalents from the grasshopper species Chothippus jucundus are held together by three (left), one (middle), and four (right) chiasmata. The middle cross-shaped bivalent is telocentric; the other two longer bivalents are submetacentric. (For an explanation of the terminology, see Fig. 12-2.) Lactopropionic orcein staining.
(Courtesy of José A. Suja and Julio S. Rufas, Universidad Autónoma de Madrid, Spain.)
Only one chiasma per pair of homolog arms is needed to hold homologous chromosomes together during meiosis I. Humans have 39 such arms on the 23 pairs of homologous chromosomes, if one excludes the five acrocentric short arms, which do not normally recombine. Remarkably, there is typically only one chiasma produced for most arms; human males typically have 46 to 53 chiasmata (Fig. 45-11).
Cohesion and Chromosomal Movements during Meiosis I
Chromosomes in mitosis achieve a dynamic alignment at metaphase as a result of a balance of forces in the spindle. The two kinetochores of the sister chromatids are attached to opposite spindle poles, and motor proteins located on the chromosomes actively pull each chromatid toward the pole that its kinetochore faces. This force does not produce any net poleward movement during metaphase because the two sister chromatids are held together by cohesion across the centromere until the onset of anaphase (see Fig. 44-16).
In meiosis I, paired homologs (called bivalents) are balanced at the metaphase plate. The structure of bivalents has two important differences from that of mitotic chromosomes. First, the kinetochore of each homolog is composed of the two kinetochores of the sister chromatids fused and acting as a single unit. The structure of the meiosis I kinetochore is most easily explained if the two kinetochores are each rotated 90 degrees toward one another relative to their position on mitotic chromosomes (Fig. 45-12A). In yeast, this coorientation of sister kinetochores requires the presence of a meiosis-specific kinetochore protein—monopolin—that associates with sister kinetochores from pachytene until anaphase of meiosis I. Monopolin recruits a protein kinase to kinetochores, but the critical kinase substrates are not known. In some organisms, a strand of material visualized by a specialized silver-staining protocol connects the sister kinetochores. This physical connection is not broken in anaphase I.
A second major difference between bivalents and mitotic chromosomes is in the force that resists the poleward pulling of the kinetochores and restrains the bivalent at the spindle midzone at metaphase. In meiosis I, this force arises from the adherence of homologs at chiasmata on the chromosome arms (Figs. 45-2 and 45-10). Thus, recombination and chiasma formation are essential parts of the mechanism that guarantees the orderly segregation of homologous chromosomes in meiotic anaphase I. However, recombination alone is not sufficient to ensure the proper segregation of bivalents at meiosis I. This is shown most clearly by the desynaptic mutant of maize. In this mutant, homologous chromosomes synapse apparently normally, and normal numbers of recombination events occur, producing chiasmata. However, these chiasmata frequently fall apart as the cells enter the first meiotic M phase. As a result, the homologs tend to segregate randomly in meiosis I. The underlying defect in the desynaptic mutation is not known, but the mutation behaves as would be expected for a defect in chromatid arm cohesion.
Work in yeasts, D. melanogaster, and Xenopus laevis, has identified a protein complex, the cohesin complex, that is required to hold sister chromatids together (see Fig. 44-16). Although this has not been proven, cohesin might form a ring that encircles sister chromatids, linking them to one another. In mitosis, cleavage of the cohesin component Scc1 is thought to open the ring, allowing sister chromatids to move apart.
Tight arm cohesion between sister chromatids causes an interesting problem for the formation of chiasmata. Presumably, when the DNA is exchanged in recombination, the protein backbone of the chromosome axis must also be exchanged. This process apparently occurs within the context of the synaptonemal complex and might involve significant topologic remodeling of the chromosomal axes.
Once chiasmata are assembled, they are held in place by cohesion between the arms of the sister chromatids (Figs. 45-7 and 45-12). This cohesion is retained throughout meiotic prophase and is released only at the onset of anaphase in meiosis I as Rec8 along the chromosome arms is cleaved. Separation of sister chromatid arms “dissolves” the chiasmata, allowing the paired homologous chromosomes to move to opposite spindle poles. In the meantime, the Rec8 at centromeres is protected from cleavage and continues to hold the sister chromatid centromeres tightly paired until anaphase of meiosis II. This protection requires a class of proteins called Shugoshins (from the Japanese, meaning “guardian spirit”), whose mechanism of action is being investigated. Shugoshin requires phosphorylation by Aurora B kinase to associate stably with centromeres and protect Rec8. This is another job for the chromosomal passenger complex (Fig. 44-10). Centromeric cohesion is released by cleavage of Rec8 at the onset of anaphase II in a process that resembles the release of cohesion during mitosis.
Behavior of the Sex Chromosomes in Meiosis
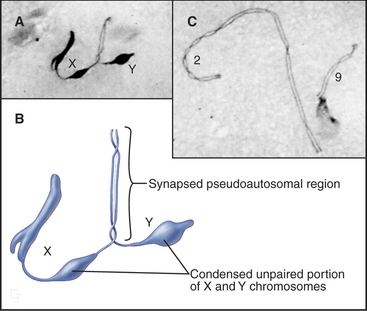
Since genetic recombination is required to stabilize homologous chromosomes at the metaphase plate in meiosis I, how is this accomplished for the X and Y chromosomes? The answer in most mammals is that the X and Y chromosomes have a short region of homologous sequence (about 2.6 million base pairs in humans) that does pair and undergo genetic recombination during meiosis. This pseudoautosomal region must undergo genetic recombination in every meiosis I cell for the X and Y chromosomes to be partitioned correctly. Thus, the X and Y chromosomes act like short homologous chromosomes with large regions of unrelated DNA attached (Fig. 45-13). Unpaired regions of the X and Y chromosomes become highly condensed during late pachytene.
Cell-Cycle Regulation of Meiotic Events
Meiosis employs the full set of functions that regulate the division of somatic cells (see Chapters 40 to 43). However, the peculiarities of the meiotic cell cycle require further mechanisms of regulation. One major difference between meiotic cells and somatic cells is that the meiotic chromosomes must undergo recombination and form chiasmata to segregate properly at the first meiotic division. Yeasts appear to lack a mechanism to detect whether recombination has occurred. Yeast cells that are completely defective in recombination proceed through meiosis with normal timing but disastrous consequences; the chromosomes fail to align properly during meiosis I, and they assort randomly to the daughter cells. On the other hand, yeast cells can detect the presence of stalled or abnormal recombination intermediates. Such intermediates accumulate if there are problems with the core recombination en-zymes or if the assembly of the synaptonemal complex (required for the completion of recombination) is defective. When such problems are detected, cells arrest late in meiotic prophase I. This has been called the pachytene checkpoint, but in fact, the cells arrest late in meiotic prophase by a mechanism that is probably analogous to the G2 DNA damage checkpoint in somatic cells. Mammalian germ cells that arrest owing to defects in recombination are eliminated by apoptosis.
The same proteins may mediate mitotic and meiotic checkpoints. Proteins such as ataxia telangiectasia–mutated (ATM) kinase, ataxia telangiectasia and Rad3-related (ATR) protein kinase, and their downstream effectors (see Fig. 40-4) are associated with meiotic chromosomes in prophase.
Suppression of DNA Replication between Meiosis I and Meiosis II
One unique aspect of meiosis is that the process involves two M phases with no intervening S phase. On exit from meiosis I, Cdk1 kinase is reactivated immediately. This blocks assembly of prereplication complexes (see Fig. 42-7), thereby blocking DNA replication. At least two pathways contribute to reactivation of Cdk1.
The first involves downregulation of translation of Wee1 protein kinase in meiosis. Wee1 is a mitotic inhibitor (see Fig. 40-14) that inactivates Cdk1 by phosphorylation at Tyr15. The absence of Wee1 in meiosis I was first observed in X. laevis but seems to be a universally conserved way of reactivating Cdk1 without an S phase. Ectopic expression of Wee1 in matureX. laevis oocytes prevents reactivation of Cdk1 immediately after the meiosis I division. As a result, the oocytes reenter interphase and replicate their DNA. Meiotic cells also express a specialized isoform of Cdc25, the phosphatase that counteracts Wee1 (see Fig. 43-1).
A second mechanism for differentiating meiosis from mitosis involves activation of a specialized mitogen-activated protein (MAP) kinase pathway (see Fig. 27-6) by c-Mos, a meiotic-specific MAP kinase kinase kinase. This pathway activates Cdk1 and other unknown substrates, with profound effects on the meiotic cell cycle (see next section).
The Metaphase II Arrest and the MAP Kinase Pathway
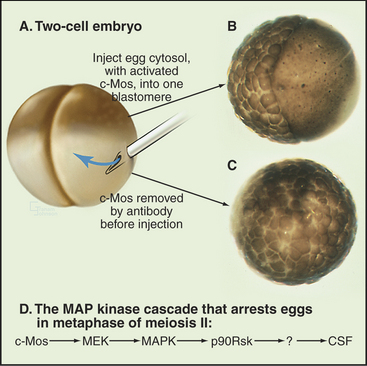
Following their activation and release from the ovary (ovulation), oocytes of many vertebrates arrest in metaphase II of meiosis until they are fertilized. The activity that is responsible for this arrest was discovered in X. laevis eggs arrested in metaphase of meiosis II and is called cytostatic factor (CSF). Injection of cytoplasm containing CSF into one blastomere of a two-cell frog embryo blocks the next cell cycle at metaphase, just like the egg (Fig. 45-14). Therefore, CSF can even block somatic cells indefinitely at metaphase in mitosis. CSF activity appears in meiosis II and disappears after fertilization.
One active component of CSF is the X. laevis homolog of a well-known viral oncogene, v-mos, the transforming gene of the Moloney murine sarcoma virus, which causes solid tumors in mice. DNA hybridization was used to isolate a corresponding cellular gene, c-mos, from X. laevis. The v-mos gene is a mutated form of the cellular c-mos gene. Vertebrates express c-mos exclusively in oocytes and eggs. Injection of either v-Mos or c-Mos proteins into dividing blastomeres of early frog embryos arrests the cells at metaphase (Fig. 45-14). These experiments led to the proposal that c-Mos was CSF.
CSF arrest requires the MAP kinase (MAPK) signal transduction pathway (see Fig. 27-5). Mos activates the pathway by phosphorylating MEK (MAPK-activating kinase), which then activates MAPK. MAPK then activates a downstream kinase called p90Rsk (Fig. 45-14D). Introduction of constitutively active c-Mos or p90Rsk into X. laevis eggs is sufficient to induce CSF arrest. However, this is not the whole story, because metaphase arrest is maintained in extracts depleted of p90Rsk. Thus the pathway must include at least one unidentified step beyond p90Rsk.
New research showed that an APC/C inhibitor called Emi2 is a critical component of CSF. A burst of cytoplasmic Ca2+ released at fertilization (see Fig. 26-15) activates protein kinase A, which phosphorylates Emi2. This modification creates a binding site for polo kinase, which then also phosphorylates Emi2. Polo phosphorylation marks Emi2 for destruction, resulting in activation of APC/C, termination of the CSF metaphase arrest, and completion of meiosis II.
Timing of Meiosis in Humans
In females, each ovary contains a total of about 100,000 primordial follicles, each with an oocyte that is arrested in the diplotene stage of meiosis at about the twelfth to sixteenth week of fetal life. Following puberty, a small number of oocytes become activated and grow each month. One of these activated oocytes matures fully and is shed in response to a surge of luteinizing hormone. The others undergo programmed cell death and degenerate in a process known as atresia. As the oocyte is shed from the ovary, it completes meiosis I and becomes arrested at metaphase of meiosis II by CSF. It remains arrested at this stage until fertilization occurs.
Meiotic Defects and Human Disease
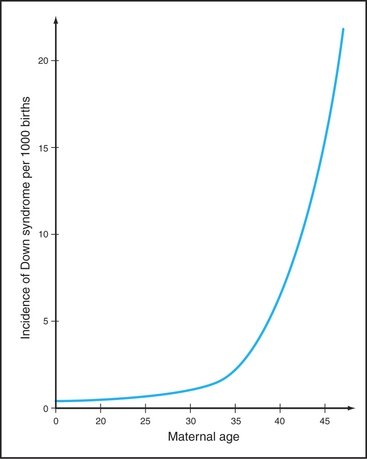
The frequency of certain types of aneuploidy, such as trisomy for chromosome 21, increases with the age of the mother. Statistics indicate that only 0.04% of children of mothers who are 20 years old have trisomy 21. This number rises dramatically with maternal age; nearly 5% of the conceptions in mothers 45 years old have trisomy 21 (Fig. 45-15). This maternal age effect is a leading cause of human genetic disease. Some believe that during the many years of arrest of oocytes in meiosis I diplotene, chiasmata joining homologous chromosomes gradually dissociate. A mechanism to explain this is the progressive loss of cohesion between sister chromatids as the mother ages. Mice with a mutation in a key subunit of the cohesin complex (Fig. 45-7; see also Fig. 13-19) exhibit a pattern of chromosome nondisjunction with increasing maternal age that looks much like that seen in aging human mothers. Of course, other factors that are required for accurate chromo-some segregation also may be lost or inactivated as oocytes age.
Not all cases of human aneuploidy are the fault of the mother. One of the most common aneuploidies, 45, X (see Table 45-1 for an explanation of nomenclature), which accounts for nearly 10% of spontaneous abortions, involves the loss of the paternal X or Y chromosome 70% to 80% of the time. In addition, about 7% of instances of trisomy 21 are of paternal origin. Clearly, more than one mechanism is responsible for the generation of aneuploid offspring.
These rather sobering statistics reveal two important facts about human reproduction. First, the production of gametes is error prone. This has been confirmed by direct studies, in which 20% of eggs and 3% to 4% of sperm were found to have chromosomal abnormalities. Second, the much lower rates of chromosomal abnormalities seen in live births (about 0.3% overall [Table 45-1]) reveal that spontaneous abortion is a highly efficient protective mechanism for the elimination of chromosomal imbalances that arise from errors in meiosis.
Harper L, Golubovskaya I, Cande WZ. A bouquet of chromosomes. J Cell Sci. 2004;117:4025-4032.
Hassold T, Hunt P. To err (meiotically) is human: The genesis of human aneuploidy. Nat Rev Genet. 2001;2:280-291.
Honigberg SM, McCarroll RM, Esposito RE. Regulatory mechanisms in meiosis. Curr Opin Cell Biol. 1993;5:219-225.
Loidl J. Coming to grips with a complex matter. A multidisciplinary approach to the synaptonemal complex. Chromosoma. 1991;100:289-292.
Petronczki M, Siomos MF, Nasmyth K. Un ménage à quatre: The molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423-440.
Rieder CL, Cole R. Chromatid cohesion during mitosis: Lessons from meiosis. J Cell Sci. 1999;112:2607-2613.
Roeder GS. Meiotic chromosomes: It takes two to tango. Genes Dev. 1997;11:2600-2621.
Sagata N. What does Mos do in oocytes and somatic cells? Bioessays. 1997;19:13-21.
Scherthan H. A bouquet makes ends meet. Nat Rev Mol Cell Biol. 2001;2:621-627.
Van Heemst D, Heyting C. Sister chromatid cohesion and recombination in meiosis. Chromosoma. 2000;109:10-26.
Villeneuve A, Hillers KJ. Whence meiosis? Cell. 2001;106:647-650.
Zickler D, Kleckner N. Meiotic chromosomes: Integrating structure and function. Annu Rev Genet. 1999;33:603-754.