Antigen-Presenting Molecules
Learning Objectives
• Explain the functions of HLAs
• Identify the major class I loci
• Identify the major class II loci
• Compare and contrast the structures of class I and II molecules
• Discuss the relationships between class I and class II molecules and CD4 and CD8 cells
• Identify the genes located in HLA class III loci
• Compare and contrast antigens binding to class I and class II molecules
• Understand the value of heterozygosity in species survival
• Explain how recombinant HLA molecules are generated
• Understand the value of allelic polymorphism in species survival
• Define a single nucleotide polymorphism (SNP)
• Restate the structural features of CD1 molecules
• Recognize the biologic function of CD1 molecules
• Define bare lymphocyte syndrome
• Explain the molecular defects associated with bare lymphocyte syndrome
Key Terms
Human leukocyte antigen (HLA) complex
Class I region
Class II region
Class III region
Haplotype
Heterozygosity
Allelic polymorphism
Single nucleotide polymorphism
Introduction
The immune system defends the host against microbial infections and mutant cells. In defense of the host, the immune system must differentiate between foreign proteins and “self-proteins.” Surface display proteins are used as markers of “self.” Early animal skin transplantation studies demonstrated that rejection or acceptance of grafts was dependent on “self” markers mapped to a gene cluster on chromosome 17. This gene cluster was called the major histocompatibility complex (MHC). In humans, the MHC is called the human leukocyte antigen (HLA) complex. The term is derived from the use of white blood cell alloantigens in tissue typing of donors and recipients prior to organ transplantation. Although HLAs are important in transfusion reactions, organ transplantation, and autoimmunity, their most important role is antigen presentation to T cells. In this role, HLA molecules control susceptibility or resistance to infection, the generation of autoimmune responses, and antibody-mediated and cell-mediated responses.
The Human Leukocyte Antigen Complex
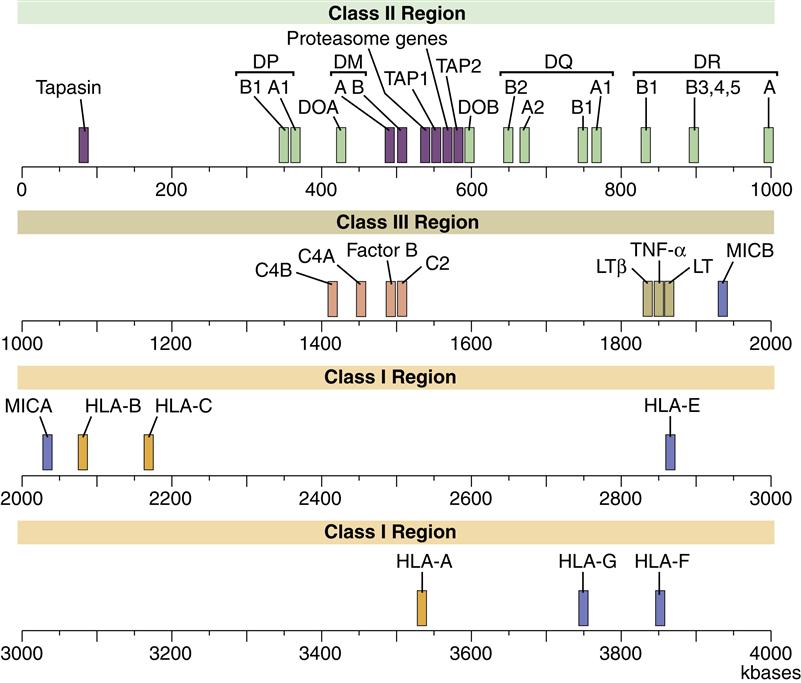
The HLA complex is a gene cluster containing 128 functional genes and 96 partial genes or gene fragments called pseudogenes. Most gene products are expressed on nucleated cells in the body, but some are secreted proteins that augment an inflammatory response. On the basis of gene product structure and function, the HLA gene complex is subdivided into class I, II, and III regions. The function of pseudogenes is unclear, but evidence suggests that pseudogenes are coding sequences that contribute to the genetic diversity of HLA molecules. A map of the human MHC or HLA complex is shown in Figure 4-1.
Class I Region
The class I region is subdivided into three major loci, termed HLA-A, HLA-B, and HLA-C, and minor loci consisting of HLA-G, HLA-E, HLA-F, HLA-H, and HLA-J and MHC class I chain–related genes (MIC). The major class I loci (A, B, and C) present antigens to CD8 T cells.
Minor loci have various immunologic functions. HLA-E interacts with CD94 and NKG2 receptors on natural killer (NK) cells to augment or inhibit NK cell function. HLA-F has an unknown function. HLA-G presents a broad range of self-polypeptides with similar molecular structure and is found in high concentrations on the surface of extravillous cytotrophoblast and the placenta. The role of HLA-G in pregnancy is unclear, but it may shift immune responses from Th1 to Th2. The functions of HLA-H and J are unclear.
The MIC family represents a nonclassic HLA gene complex. Gene products are expressed as a stress response to virus infection or cellular damage in the intestine and synovia. Their structure resembles a class I molecule, but MIC gene products cannot present antigens. Expression of MIC marks the cell for destruction by CD8 and NK cells.
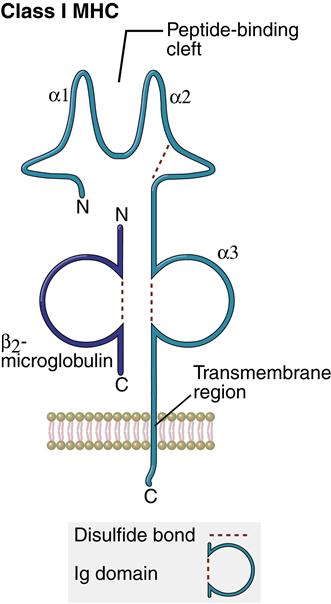
Structure of Class I Molecules
Class I molecules comprise a single glycosylated α-chain (394 amino acids), which is noncovalently bound to β2-microglobulin (94 amino acids). The class I protein consists of two intracellular and three extracellular domains. The two intracellular domains attach the molecules to the cell membrane and extend into the cytoplasm. The three extracellular domains—designated α1, α2, and α3—are each composed of 90 amino acids. The α1– and α2– domains have unique helical structures that form an antigen-binding cleft used to present antigens to T cells (Figure 4-2).
Peptides are tethered to the ends of the binding groove usually at positions 2 and 9. Thus, the binding cleft accommodates only small peptides containing 8 to 12 amino acids. Flexing of unbound peptides and side chains within the binding cleft creates a three-dimensional epitope structure that is recognized by T cells. The α3-domain stabilizes the interaction between class I molecules and the lymphocyte T cell receptor.
Class II Region
Class II gene products are induced and expressed on monocytes, macrophages, dendritic cells, and B cells. Within the class II region are three loci (DP, DR, and DQ) involved in antigen presentation. Class II molecules present antigens to CD4Th1 and CD4Th2 cells. Other genes in the class II region code for proteins that are important in antigen processing.
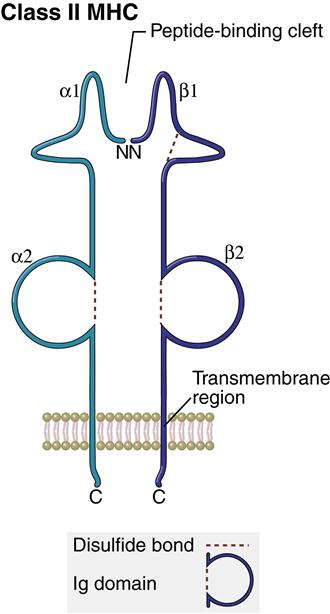
Structure of Class II Molecules
The class II region has three major loci (DP, DQ, and DR). Class II molecules consist of heavily glycosylated α- and β-chains. Although both chains have similar structure, the α-chain is larger (30–34 kiloDaltons [kDa]) compared with the β-chain (26–29 kDa). An antigen-binding cleft comprises the α1– and β1-domains (Figure 4-3). The peptide binding to the class II molecule occurs in the middle of the binding cleft at positions 1, 4, 6, and 9. Peptides are bound in a manner analogous to a long pipe being held in a centrally located vise. Peptides are held in place by hydrogen bonding and van der Waals forces. All class II antigens bind at the same anchor positions. However, they are much larger than class I antigens (10–30 amino acids) and protrude from either end of the cleft.
Class III Region
The class III region has 62 genes, which have diverse functions. Most class III gene products are not membrane proteins but are secreted into the environment. Products include three complement components and the tumor necrosis factor (TNF).
Inheritance of Human Leukocyte Antigens
A set of HLAs (HLA A, B, C, DP, DQ, and DR) on the same chromosome is called a haplotype. The genotype of each individual consists of two haplotypes. One set of HLA genes (N=6) is inherited from the father, and the other haplotype (N=6) is inherited from the mother. Both haplotypes are co-dominant and expressed on cells. HLA genes usually are inherited in blocks as haplotypes. Occasionally, deoxyribonucleic acid (DNA) segments are exchanged in a process called gene conversion. During meiosis, two homologous chromosomes with multiple genes arranged in tandem are misaligned. Crossing over of parental haplotypes and DNA recombination allows individual genes or parts of genes to transfer from one chromosome to another without any loss of function. Exchange of DNA causes multiple amino acid changes in the original gene and the formation of a recombinant HLA molecule.
Heterozygosity
The inheritance of duplicate genes at the same locus may be the result of a population balancing selection process, which, in theory, provides a survival advantage to heterozygotes. All HLA molecules can present antigens to T cells, but each HLA molecule binds a different range of antigens. In effect, the inheritance of paternal and maternal haplotypes doubles the antigen-presenting capability of the host and increases the probability that most individuals within a species will be heterozygotes. Heterozygotes at an HLA locus are more resistant to disease than are homozygotes because they have a more varied repertoire of antigen-presenting HLA molecules.
Allelic Polymorphism
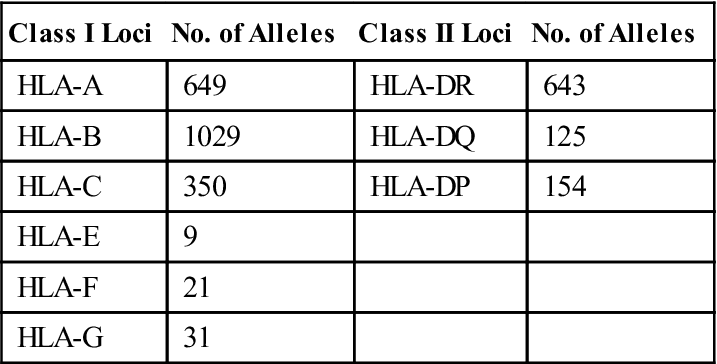
Additional genetic diversity critical to the disease resistance is provided by allelic and single nucleotide polymorphisms (SNPs). When several alternate forms of the same gene are present, the gene is termed polymorphic. Each variant of a polymorphic gene is called an allele. Over 3000 different alleles are present within the HLA complex. Two thousand polymorphic alleles have been reported in class I loci alone. Over 900 different alleles also are found in class II DR, DQ, and DP loci. Nonclassic, minor loci such as HLA-F, HLA-G, HLA-H, and HLA-J are less polymorphic compared with antigen-presenting loci and have restricted tissue distribution. The different alleles present in major loci are shown in Table 4-1. SNPs are single base pair changes in genes coding for each HLA allele. In effect, SNPs create additional alternative forms of each allele.
Table 4-1
< ?comst?>
| Class I Loci | No. of Alleles | Class II Loci | No. of Alleles |
| HLA-A | 649 | HLA-DR | 643 |
| HLA-B | 1029 | HLA-DQ | 125 |
| HLA-C | 350 | HLA-DP | 154 |
| HLA-E | 9 | ||
| HLA-F | 21 | ||
| HLA-G | 31 |
< ?comen?>< ?comst1?>
< ?comst1?>
< ?comen1?>
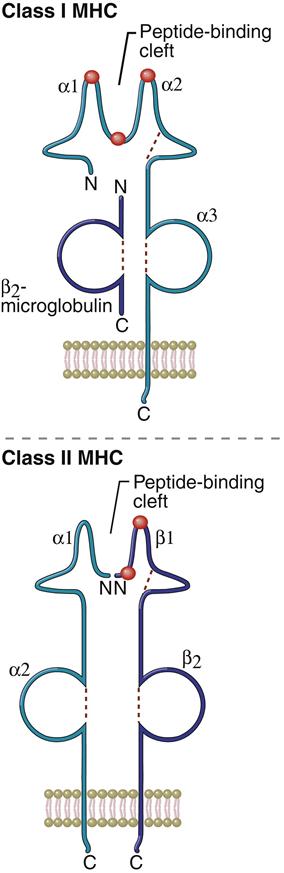
Allelic polymorphism and SNPs are associated with nonsynchronous amino acid substitutions in the α-helical sides of the binding cleft or alterations in the peptide anchoring β-strands forming the floor of the cleft. A change in the binding cleft alters the pattern of antigens binding to the allele (Figure 4-4).
Allelic polymorphism is a response to the constant and continuing evolution of microbes and is essential to the survival of the species. Within an HLA locus, the presence of different alleles presenting a wide range of antigens from the same microbe ensures that some members of a species will survive the microbial infection. In addition, allelic polymorphism protects some members of a species when a microbe inadvertently expresses a specific HLA allele. Microbes that express an HLA allele would be considered “self” and evade immune detection. As a consequence of infection, significant population mortality may occur among individuals carrying the allele. However, individuals expressing other HLA polymorphic alleles within the same loci would be unaffected.
Human Leucocyte Antigen Single Nucleotide Polymorphisms and Disease
The HLA molecule’s ability or inability to present antigens to T cells determines an individual’s resistance or susceptibility to microbial infections. For example, self-limiting hepatitis B is associated with the expression of HLA-DR13. Individuals homozygous for DR13 have more efficient presentation of hepatitis antigens, a strong vigorous CD4 response, and accelerated viral clearance. Chronic hepatitis B infections develop in individuals expressing other HLA DR molecules. HLA alleles also control susceptibility to infections. The strongest relationship between HLA and disease is found in HLA alleles containing SNPs. For example, individuals expressing (DRB1∗1501) molecules have a high risk for developing pulmonary tuberculosis. It is assumed that these class II molecules cannot present mycobacterium antigens to CD4Th1 cells.
Antigen-Presenting Molecules Outside the Human Leucocyte Antigen Complex
The CD1 family of genes is involved in antigen presentation. Unlike HLA molecules, they are encoded by a gene cluster on chromosome 1. The configuration of the CD1 molecule is similar to a class I protein with respect to subunit organization and the presence of B2-microglobulin. CD1 molecules present highly conserved glycolipids to CD4Th1 cells. Glycolipids include mycolic acid, glucose monomycolate, phosphoinositol mannosides, and lipoarabinomannan derived from the Mycobacterium species that cause tuberculosis and leprosy.
Two families of CD1 molecules exist: (1) Group I consists of CD1a, CD1b, and CD1c. (2) CD1d is the only member of group II (Table 4-2). CD1a is expressed on thymocytes, dendritic cells, and Langerhans cells. CD1b is expressed on monocytes and macrophages. CD1c is found on circulating B cells, the splenic mantle zone, and tonsillar B cells. Monocytes, macrophages, dendritic cells, B cells, and some nonlymphoid cells express CD1d.
Table 4-2
Tissue Distribution of Group I CD1 Molecules
| CD1 Family Member | Tissue Distribution |
| CD1a | Thymocytes, dendritic cells, Langerhans cells |
| CD1b | Macrophages |
| CD1c | Circulating B cells, splenic mantle zone and tonsillar B cells |
| CD1d | Monocytes, macrophages, dendritic cells, B cells |
It is unclear whether the five members of the CD1 family present the same antigens or different antigens. It is conceivable that each represents a redundant system for the identification of conserved molecules. Some evidence suggests that group I CD1 is promiscuous and presents an overlapping set of glycopeptides. Other CD1 molecules may have individual binding properties.
Antigen-Presenting Molecules and Immunodeficiency
Bare Lymphocyte Syndrome
The bare lymphocyte syndrome (BLS) is a form of severe combined immunodeficiency (SCID). In this syndrome, individuals lack class II molecules on the surface of B cells and monocytes. Class II molecules are not produced because of mutations in the consensus sequences of gene promoters that regulate HLA class II structural genes. Mutations prevent the docking of regulator factor X (RFX) and class II transactivator (CIIT) with the promoter genes. CIIT regulates the expression of the class II molecule.
Children with BLS have repeated infections with Candida albicans or Pneumocystis jiroveci. Common childhood viral infections with respiratory syncytial virus (RSV) or cytomegalovirus (CMV) are invariably fatal.
Treatment of Bare Lymphocyte Syndrome
Without hematopoietic stem cell transplantation, infants with this syndrome die within the first year of life. Aggressive antibiotic therapy is indicated for suspected microbial and viral infections. Agents used in the treatment of BLS are shown in Table 4-3.
Table 4-3
| Agent | Infection |
| Acyclovir | Herpes simplex, cytomegalovirus, varicella zoster |
| Fluconazole | Mucosal Candida albicans |
| Amphotericin B | Invasive Candida albicans and Aspergillus |
| Itraconazole | Aspergillus |
Summary
• Three major class I and class II loci are present within the coding regions.
• Major loci within the class I region produce glycoproteins that present antigens to CD8 cells.
• Major loci within the class II region produce glycoproteins that present antigens to CD4 cells.
• Regions outside the HLA complex code for molecules that present highly conserved molecules.