Antibody Diversity
Learning Objectives
• Recognize the three theories that explain antibody diversity
• Compare and contrast genes that code for light chains and heavy chains
• Recognize the key features of combinatorial diversity
• Explain the biologic function of complementarity regions 1, 2, and 3
• Discuss the role of the recognition signal sequence (RSS) in recombination
• Relate the biologic significance of the 12/23 rule in the recombination of V(D)J genes
• Understand the role of RAG-1 and RAG-2 in recombination
• Identify the role of the Artemis enzyme in recombination
• Understand how messenger ribonucleic acid (mRNA) slicing attaches C regions to VJ or VDJ
• Relate junctional diversity to the antigen-combining pocket
• Identify the definition of allelic exclusion
• Explain the role of affinity maturation in immune response
Key Terms
Affinity maturation
Allelic exclusion
Artemis
Combinatorial diversity
Deoxynucleotidyl transferase
Junctional diversity
Ligase
Messenger ribonucleic acid (mRNA) splicing
Nonhomologous end joining
One turn–two turn rule
P-nucleotides and N-nucleotides
RAG enzymes
Recognition signal sequences
Somatic mutation
Signal joints
Introduction
Three theories have been put forth to explain antibody diversity, which allows B cells to generate an antibody repertoire capable of reacting with a wide range of antigens: (1) The germ-line theory postulates that separate genes exist for each antibody molecule and that the antibody repertoire is largely inherited. (2) The deoxyribonucleic acid (DNA) rearrangement theory proposes that a limited number of genes undergo genetic rearrangements to create antibody populations. (3) Finally, the somatic mutation theory proposes that a limited number of inherited genes undergo mutations to general antibody repertoires. In vivo and in vitro studies have demonstrated that both the DNA rearrangement theory and the somatic mutation theory provide the most plausible explanations for antibody diversity.
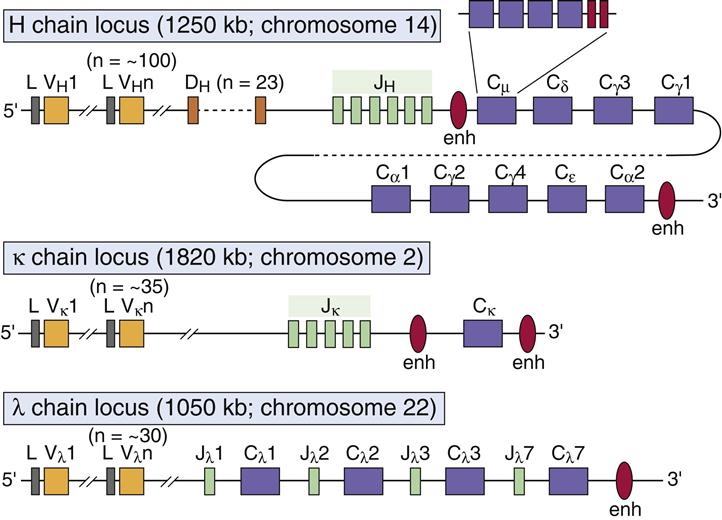
Antibodies are encoded by different germ-line genetic loci. Variable (V) region, joining (J) region, and constant (C) region gene products are assembled into a functional antibody. Variable portion genes (V) code for amino acids that constitute the framework regions of the variable region, and three hypervariable complementarity-determining regions (CDR1, CDR2, and CDR3). The hypervariable regions form the three-dimensional antigen-binding pocket. Antibody specificity is determined by the specific amino acid sequences in CDR3. The joining (J) segment is, in reality, part of the V region and provides some of the framework for the antigen-binding pocket. Only heavy chains have an additional diversity (D) gene.
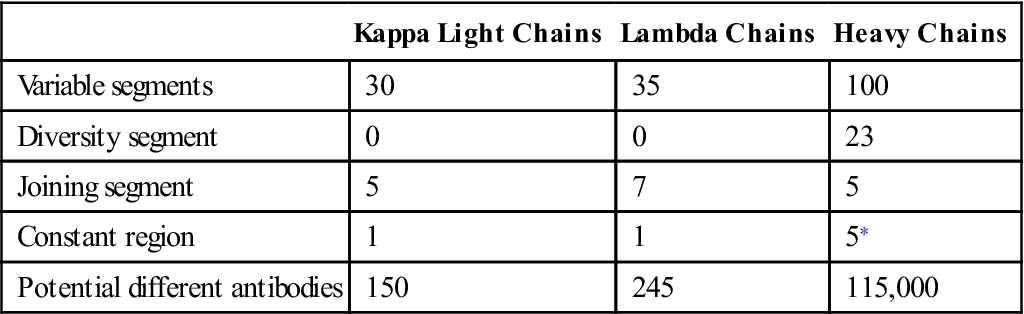
Antibody diversity is generated from the large number of V, J, D, and C genes available for recombination. Light-chain loci have 30 to 35 genes encoding for the variable (VL) regions (Table 10-1). Five to seven genes code for JL segments in kappa (κ) or lambda (λ) light chains, respectively. Lambda and kappa light chains have one highly conserved constant region.
Table 10-1
Number of Genes Coding for V, D, J, and C Regions in Light and Heavy Chains
< ?comst?>
| Kappa Light Chains | Lambda Chains | Heavy Chains | |
| Variable segments | 30 | 35 | 100 |
| Diversity segment | 0 | 0 | 23 |
| Joining segment | 5 | 7 | 5 |
| Constant region | 1 | 1 | 5∗ |
| Potential different antibodies | 150 | 245 | 115,000 |
< ?comen?>< ?comst1?>
< ?comst1?>
< ?comen1?>
< ?comst1?>< ?comen1?>∗< ?comst1?>< ?comen1?>Constant region for immunoglobulin G (IgG), IgM, IgA, IgD, and IgE.
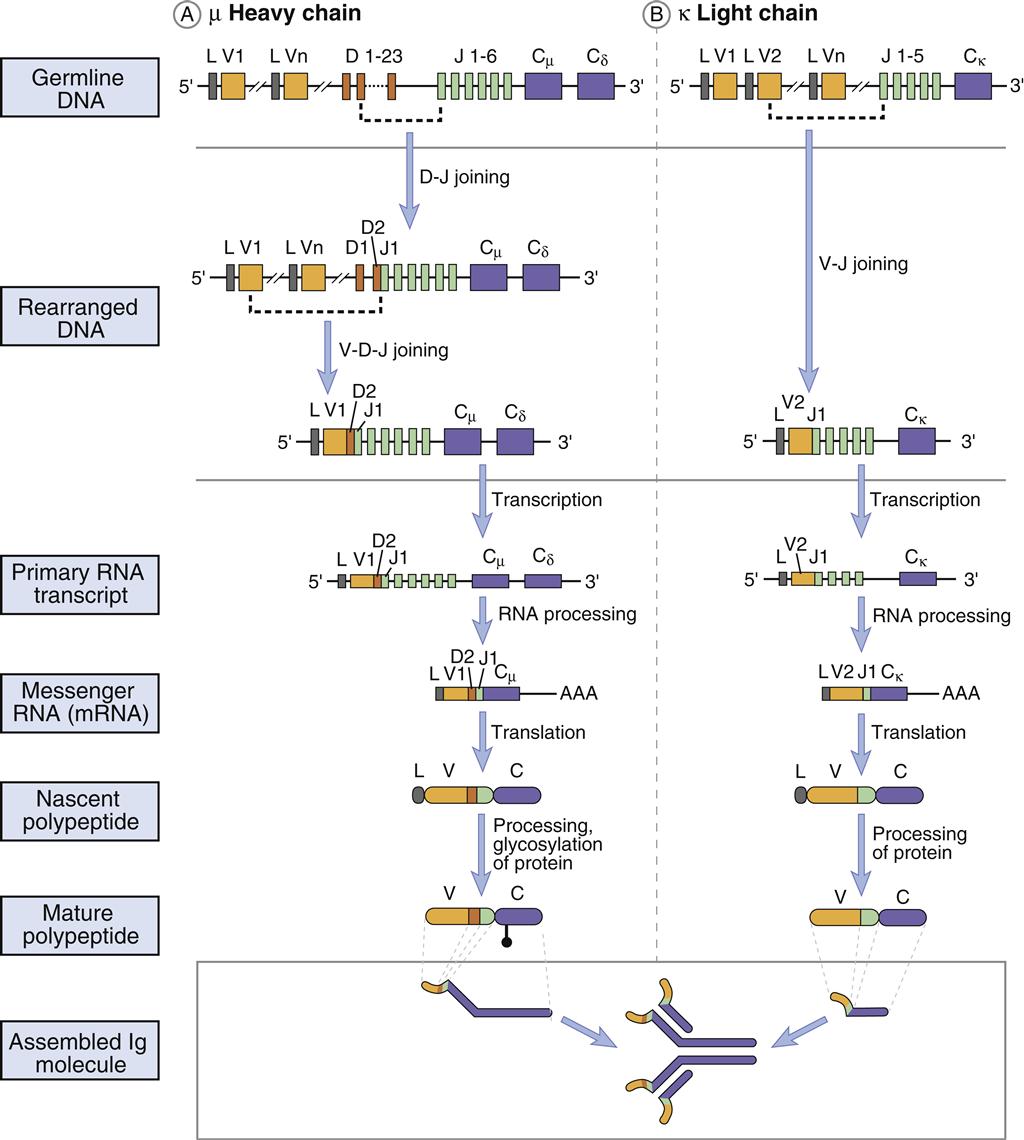
Heavy chains are larger than the light chains. A hundred genes code for heavy-chain variable (VH) regions. Diversity (D) genes (N=23) are inserted between V and J genes. J genes (N=5) are linked to the constant region (N=5). The constant region may be from one of the five antibody isotypes (mu [μ], gamma [γ], alpha [α], epsilon [ε], or delta [δ]). An assembled heavy chain consists of VJDC gene products (Figure 10-1).
Combinatorial Diversity
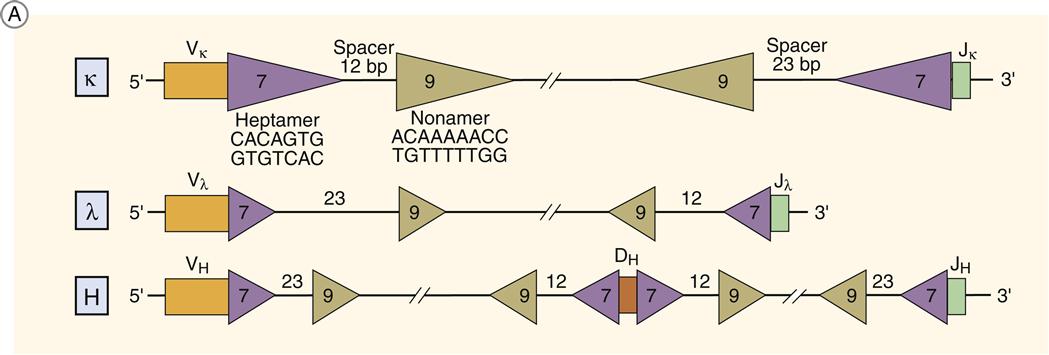
The rearrangement of genes takes advantage of recognition signal sequences (RSS) flanking the V, J, and D sequences. The RSSs consist of a heptamer (5¢CACAGTG-3¢) followed by either 12 bases (RSS 12) or 23 bases (RSS 23) and an AT-rich nonamer (5¢-ACAAAAACC-3¢). Genes can only recombine when they are located on the same side of the chromosome. RSSs align genes on the same side of the DNA because the spacer placement also corresponds to one (12 AA) or two turns (23 AA) of DNA. This recombination restriction is called the 12/23 rule or the one turn–two turn rule. Only RSS 12 and RSS 23 segments can combine.
Light Chain VJ Recombination
The two types of antibody light chains are called lambda and kappa light chains. A single antibody can have either two kappa light chains or two lambda light chains, but not both. Recombination of light chain V and J provides the simplest example of recombination. VL RSSs have a 23-base spacer on the 3¢ end. Joining genes have a 12-base RSS on the 5′ end.
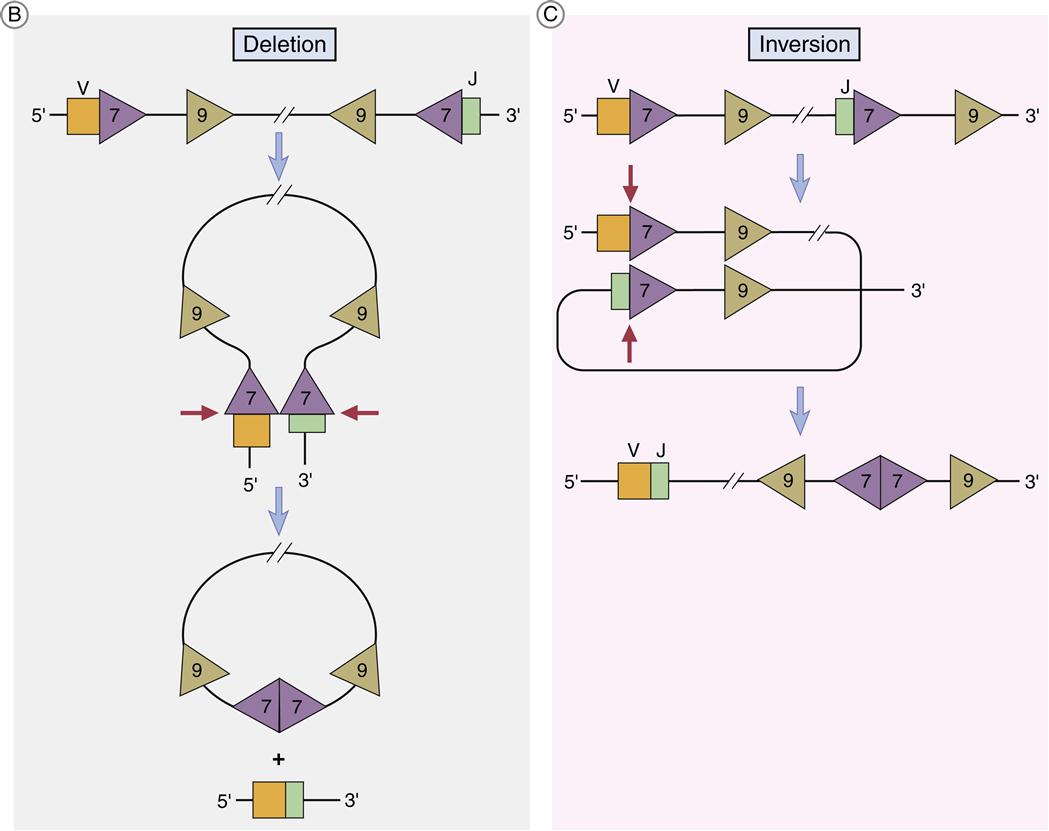
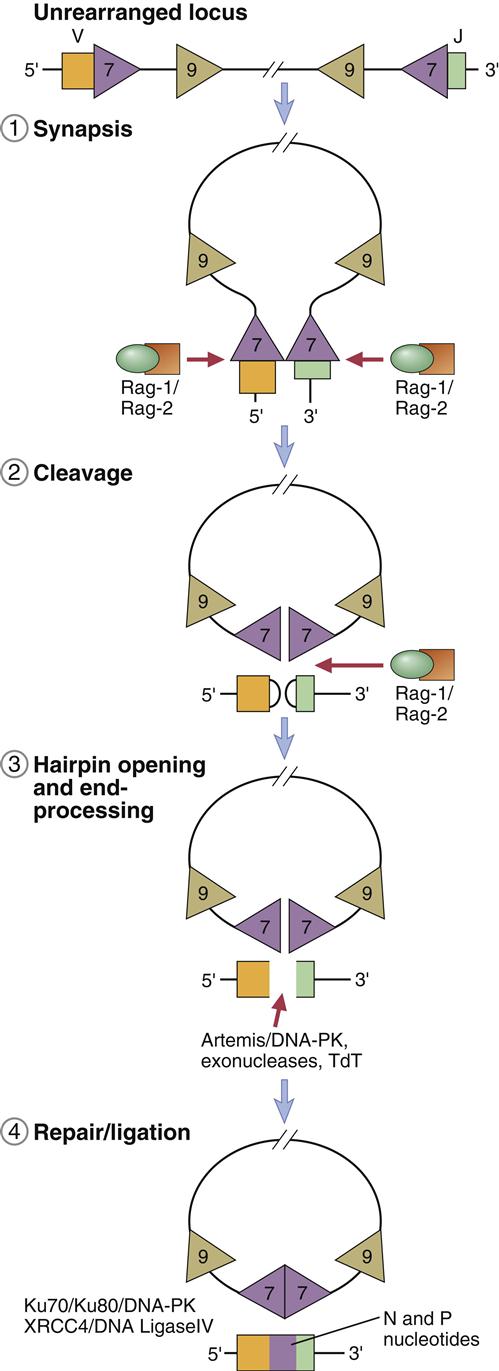
Light chain V and J coding genes or exons are separated by a single intron, and the assembly of the light chain requires only one recombination event. RAG-1 and RAG-2 enzymes, which are part of a recombinase family, facilitate the recombination of genes. Tetrameric complexes of RAG-1 and RAG-2 bind to 23-base RSSs on a VL gene and a 12-base spacer on the JL gene and bring the genes close. As the genes move closer, a large loop, called a signal joint, is created. The loop contains introns located between the two coding genes. As the signal joint is deleted from the chromosome, RAG-1 nicks one strand of the double-stranded DNA between the coding gene and the RSS heptamer. The 3¢OH group from the nicked strand reacts with 5¢OH on the same strand to form a hairpin. RAG-1 then attacks the second strand and forms another strand-specific hairpin curve (Figure 10-2).
The hairpins must be removed from the coding ends before V and J genes can be ligated. The opening of the hairpins is facilitated by a nuclease called Artemis and DNA-dependent kinase (DNA-PK). Using ligase IV, the double strands are tied together by using a specialized process called nonhomologous end joining (NHEJ). The process is nonhomologous because strand breaks are ligated without requiring a template.
In 50% of the kappa chains, V genes have a reverse orientation and transcriptional direction. In this case, looping inverts the DNA to align the V and J segments. Recombination still occurs at the heptamer regions, but the RSSs are retained in the chromosome in an inverted orientation (Figures 10-2 and 10-3).
Heavy Chain VDJ Recombination
Heavy chains have a complex RSS system. VH genes have a 23-base spacer on the 5¢ end, and J genes have a similar base pair spacer on the 3¢ end. The genes will not recombine until a diversity gene is inserted between V and J genes. Diversity genes have 12 base-pair RSSs at both the 3¢ and 5¢ ends, which allows binding to both V and J genes. The partially assembled heavy chain consists of VDJ genes.
Attachment of Constant Regions
When VJ or VDJ genes are ligated, a promoter in the V region initiates transcription of an mRNA that contains introns between J and C genes. A process called mRNA splicing is used to remove introns. After transcription of mRNA, specialized small nuclear RNA (snRNA) and small nuclear riboprotein (snRAP) combine to form a stable complex. The first three base pairs of the snRNA attach to complementary sequences in the intron. The 3¢ and 5¢ ends of the intron are brought together to form a spliceosome, or extended loop. Cleavage of the spliceosome at the intron removes the intron separating J and C genes. Following removal of the intron, the coding units are ligated. A summary of the recombination and expression of light chains and heavy chains is provided in Figure 10-4.
Junctional Diversity
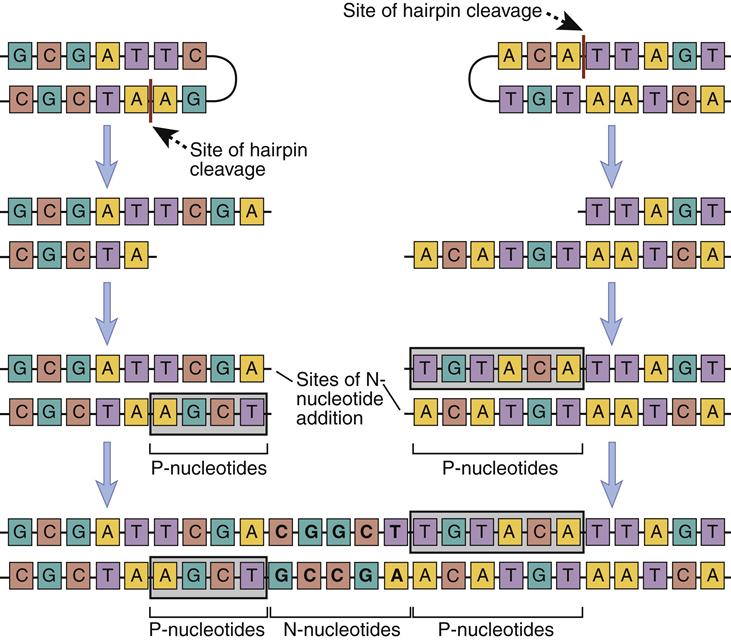
Junctional diversity occurs at the junction of the V and J segments. This region codes for the hypervariable CR3 region in the antigen-combining pocket. Changes in the amino acid sequence change the specificity of the antibody. The joining of gene V(D)J is often imprecise, and nucleotides can be lost from the ends of each gene segment. Diversity is increased by the addition of P-nucleotides and N-nucleotides. P-nucleotides are added because Artemis cleavage is often asymmetrical and creates one long DNA strand and one short DNA strand. For proper ligation, the short strand must be extended by the addition of P-nucleotides to the length of the long strand. A random addition of 2 to 20 base pairs, which are called N-nucleotides, also occurs. The addition of N-nucleotides changes the amino acid sequence in the hypervariable CR3 region. Addition of P-nucleotides and N-nucleotides is mediated by a unique enzyme called terminal deoxynucleotidyl transferase (Figure 10-5).
Nucleotides also can be removed from J genes during recombination. Following the cleavage of hairpin turns, endonucleases may inadvertently remove sequences from the exon.
Addition or removal of nucleotides often disrupts the open reading frames and the structure of the antibody. Over 66% of rearrangements in the J regions results in nonfunctional antibodies.
Summary of Mechanisms Involved in the Generation of Diversity
For the host to survive, antibodies must be able to recognize 1013 different antigens. Antibody diversity is generated by a number of different mechanisms that include combinatorial diversity and junctional diversity, insertion and deletion of random nucleotides, and somatic mutations. The product of these V(D)J recombinations generates 2×109 antibodies with different specificities. P-nucleotide and N-nucleotide additions and somatic mutations account for 1×104 different antibodies (Box 10-1).
Alleleic Exclusion
Since both maternal and paternal V(D)J and C genes are inherited, an antibody-producing B cell is diploid. Genes from only one parent (heavy) are rearranged to produce antibodies. H-chain genes from the other parent are excluded or not productively rearranged. Allelic exclusion is the term used to describe antibody production from a single rearranged set of parental genes.
Affinity Maturation
Somatic hypermutation of V genes occurs in B cells in germinal centers. In V genes, “hotspots” in the hypervariable region are present and are easily mutated. Mutations in the V region CDRs create IgM B cell receptors (BCRs) with increased affinity for antigen.
During antigen-driven B cell expansion, these cells are preferentially stimulated to produce high-affinity antibodies. This process is termed affinity maturation.
Summary