Antibodies
Learning Objectives
• Describe the basic structure of antibodies
• Discuss the function of the variable and hypervariable portion of antibodies
• Identify the functions of the C H1–C H3 regions
• Recognize the definition of the complementarity-determining regions (CDRs)
• Compare and contrast antibody affinity and avidity
• Compare and contrast the structure and antigen-binding capability of Fab and F(ab’)2
• Describe the usefulness of Fab fragments in treating drug overdoses
• Compare and contrast the structure and function of immunoglobulin M (IgM) and IgG
• Identify the structure and biologic functions of IgG subclasses
• Identify the clinical implications of IgG subclasses
• Compare and contrast the structure and function of IgA and secretory IgA (sIgA)
• Understand the mechanism used to transport IgA to the exterior
• Understand the role of mucosal-associated lymphoid tissue (MALT) and sIgA in vaccination
• Relate the structure and function of IgD
• Restate the biologic function of IgE
• Identify the definition of an anamnestic response
• Understand the mechanisms involved in the generation of an anamnestic response
• Understand the role of cytokines in isotypic switching
• Discuss the immunologic defects in transient hypogammaglobulinemia of infancy (THI)
• Discuss the immunologic defects associated with common variable immunodeficiency (CVID)
• Identify symptoms and immunologic defects associated with hyperimmunoglobulinemia E (Job syndrome)
• Describe the drugs used to treat Job syndrome
• Understand the pathophysiology of multiple myeloma
• Describe Bence Jones proteins
• Identify the drugs used to treat multiple myeloma
• Understand the pathophysiology of Waldenström’s macroglobulinemia
Key Terms
Affinity
Avidity
Anamnestic response
Bence Jones proteins
Complementarity-determining regions (CDRs)
Fab
F(ab’)2
Fc
IgM
IgG
IgA
IgD
IgE
Isotypes
Multiple myeloma
Waldenström’s macroglobulinemia
Introduction
Antibodies are soluble proteins produced by plasma cells. They are normally found in peripheral blood and external body fluids such as saliva, tears, and colostrums. Antibodies neutralize viruses or mark antigens or microbes for destruction by phagocytosis or complement lysis. A number of different synonyms for antibodies exist. When serum is placed in an electrical field, blood proteins migrate at different rates, depending on size and charge. Small-molecular-weight albumins migrate rapidly, whereas globulin fractions migrate more slowly. Antibodies are localized in the slow-migrating gamma (γ) fractions and are termed gammaglobulins. Ultra-centrifugation also can be used to separate immunoglobulins on the basis of size. On the basis of the sedimentation rate (Svedberg units) in a centrifugal field, antibodies are divided into three different molecular weights: (1) the 7S antibody fraction with a molecular weight of 150,000, (2) the 11S fraction with a molecular weight of 300,000, and (3) the 19S fraction with a molecular weight of 900,000.
Antibody Structure
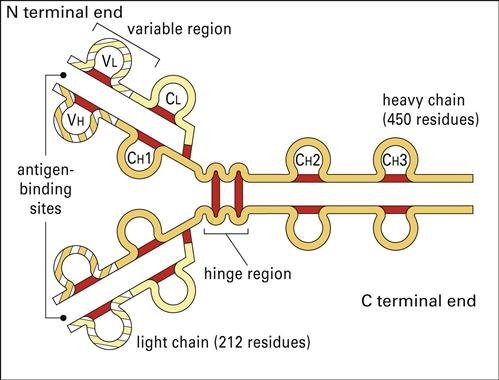
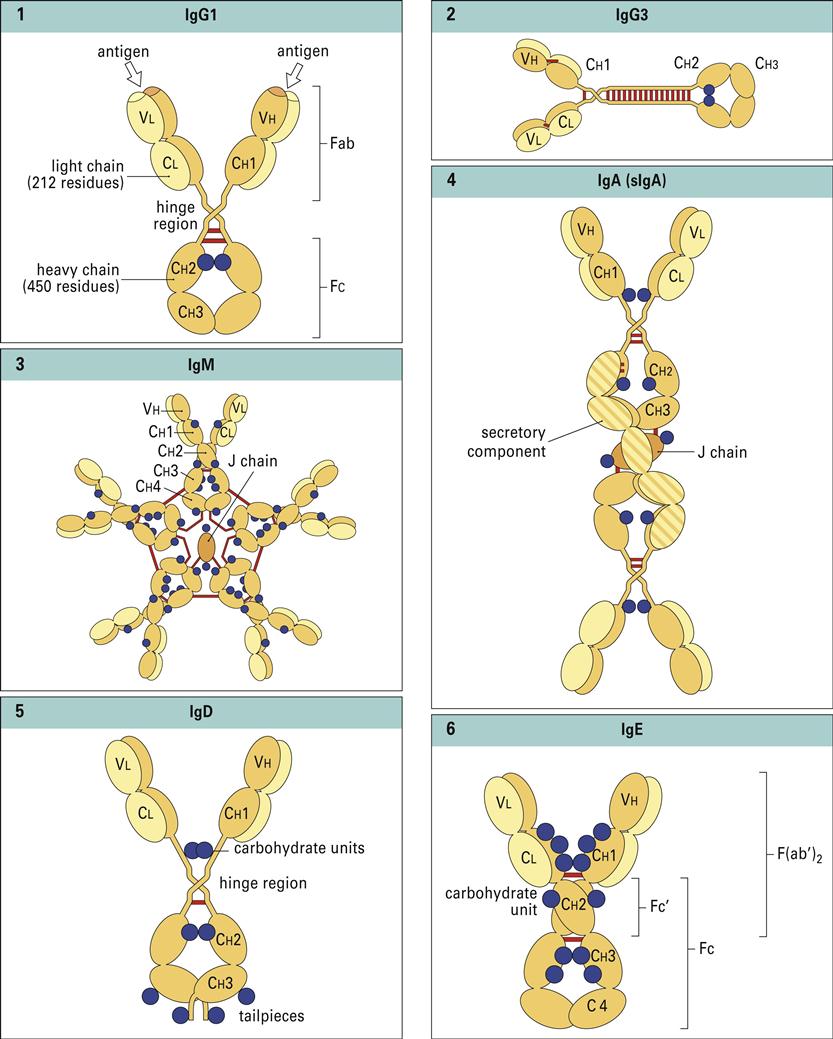
Antibodies have a basic structure which consists of pairs of heavy (H) and light (L) chains. Each heavy chain has a molecular weight of 50,000 Daltons. The weight of light chains is approximately 25,000 Daltons. Disulfide bonds link heavy chains and attach light chains to heavy chains. Heavy and light chains consist of several different globular domains that have constant or variable amino acid sequences (Figure 9-1).
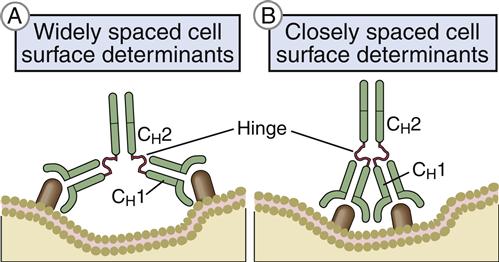
Heavy-chain constant domains are called constant regions (CH1, CH2, and CH3). Heavy-chain CH1 segments are linked to the variable (VH) domain, which is part of the antigen-combining site. The CH2 region is the hinge region that ensures antibody flexibility—the larger the hinge region, the more flexible is the antibody (Figure 9-2). Flexibility is essential because epitopes are often widely spaced on protein molecules or microbes. Attachment to cellular receptors is facilitated by the antibody CH3 region. However, some antibody subpopulations express an additional heavy-chain region (CH4) that restricts binding to select cells.
Light chains attached to the heavy chains also have constant (CL) and variable region (VL) domains. The VL domain contributes to antigen binding. These 24-kiloDalton (kDal) light chains are covalently linked to heavy chains via disulfide bonds.
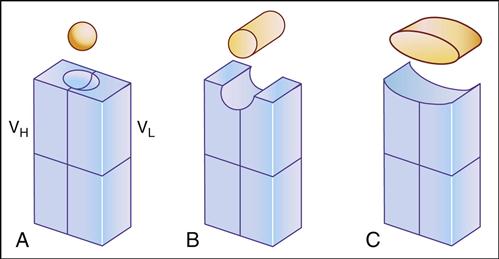
Antibody specificity is determined by the association of V regions from both heavy and light chains. In essence, V segments form a three-dimensional pocket that is the mirror image of the antigen that elicited its production (Figure 9-3).
Within the antigen-binding pocket, small 10-amino-acid hypervariable regions determine specificity at the molecular level. Amino acid sequences that form the three-dimensional antigen pocket are termed complementarity-determining regions (CDRs). Each heavy- and light-chain variable region contains three CDRs.
Affinity and Avidity
Antibody binding is described in terms of affinity and avidity. The binding strength or affinity is the result of the interaction between an antibody and a single antigenic determinant. From a practical perspective, antibody affinity is important in determining the rate at which an infection is terminated. Antibodies with high affinity will tightly bind lower concentrations of microbes and quickly terminate an infection. Low-affinity antibodies are most efficient when large antigen concentrations are present, which means that it takes longer to terminate the infections. Antibody avidity is dependent on the number of antibody-combining sites and the number of epitopes in a single antigen. It is the combined strength of multiple interactions between antibodies and epitopes. Avidity will always be geometrically higher than the affinity.
Antibody Fragments
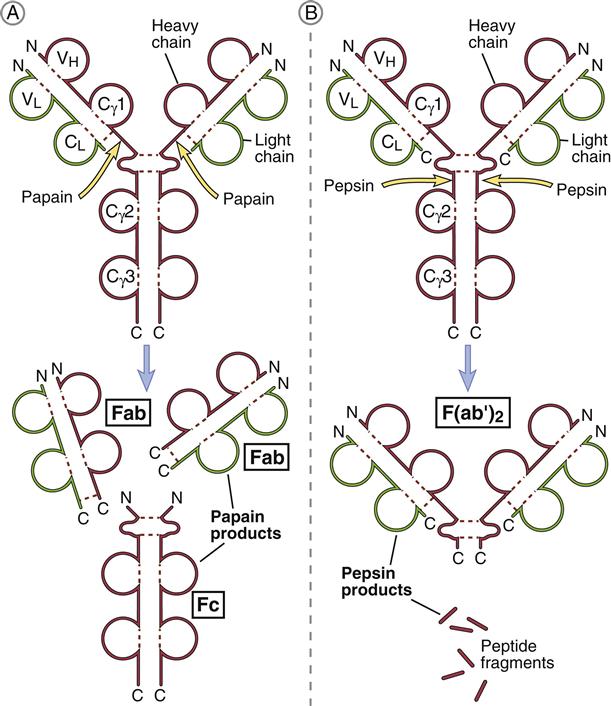
Porter delineated antibody structure by digesting antibodies with proteolytic papain and pepsin. Papain cleaves the heavy-chain CH2 domains above the disulfide bonds that connect heavy chains and yields three different fragments. The resultant pieces are identical in structure and consist of constant and variable regions of heavy (VH and CH1) and light (VL and CL) chains linked by disulfide bonds. These two fragments, each having a molecular weight of 60,000, are called Fab (fragment antigen binding). Each Fab binds a single antigen. The remaining fragment, which consists of CH2 and CH3 heavy-chain units, is easily crystallized and is called Fc (fraction crystallized). Later studies showed that the Fc portion of antibodies binds to receptors (FcR) on immunocompetent cells (Figure 9-4).
Pepsin cleaves IgG heavy chains at a point below the disulfide bond that links two heavy chains. The resulting fragment consists of two Fab linked together by heavy-chain disulfide bonds. The large-molecular-weight fragments are called F(ab’)2. Each F(ab’)2 can bind two antigens. The remainder of the antibody is reduced to small peptide fragments that have no biologic function.
Clinical Usefulness of Fab Fragments
Fab fragments are used in the clinic to treat select drug overdoses and envenomations. For example, Fab fragments are commonly used to treat digoxin overdoses or North American Crotalidae (rattlesnake) envenomations. Fab fragments neutralize the drug or the venom in blood, and the resulting equilibrium shifts away from target cell binding. Since the molecular weight of the antigen–Fab complexes are usually less than 65,000 molecular weight (MW), they pass through the kidneys and are eliminated in urine. Patients usually begin to recover within 30 minutes of receiving a bolus of Fab fragments.
Antibody Isotypes
The five antibody isotypes (IgM, IgG, IgA, IgD, and IgE) are differentiated by the chemical structure of the heavy chain. Antibody isotypes have different molecular structures, have different affinity constants, appear at different times in an immune response, and have different functions (Figure 9-5).
Immunoglobulin M Isotype
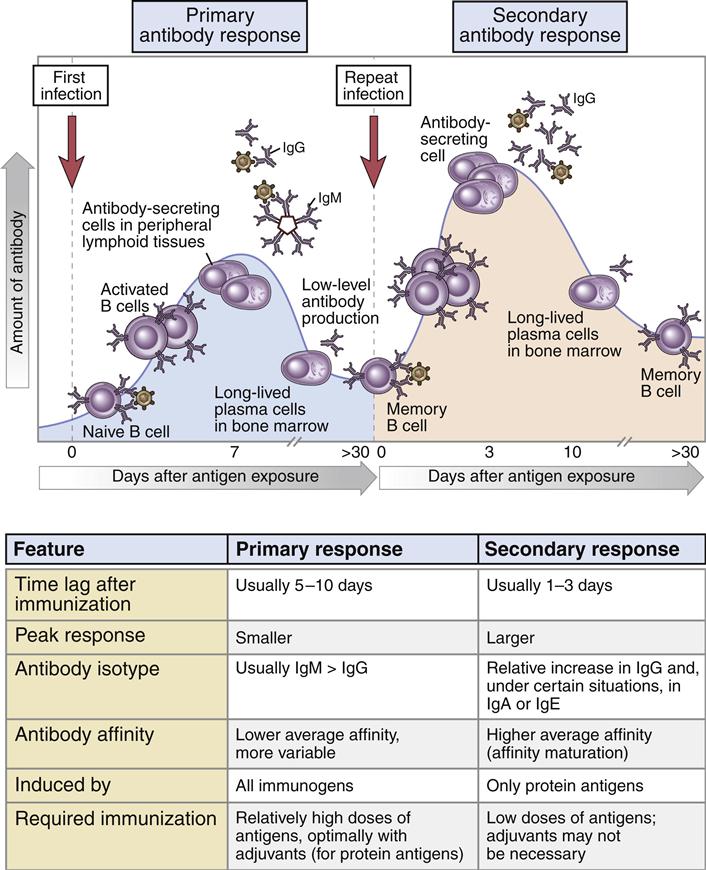
Immunoglobulin M (IgM, or 19S) is the antibody formed during the initial response to an antigen or microbe. Following antigen stimulation, IgM-producing plasma cells migrate from the lymph node to bone marrow, where they become long-lived plasma cells. A 5- to 10-day lag time occurs before IgM antibodies appear in blood, and peak IgM levels occur at 21 days. During the response, memory B cells are also produced, and they remain in the lymph node germinal centers or recirculate between the lymph node and the spleen.
IgM has the basic heavy- or light-chain antibody structure but exists in pentameric, secretory, and monomeric configurations. In the pentameric configuration, five antibody units are covalently linked by disulfide bonds at adjacent CH3 domains. An additional molecule, called a J-chain, also is attached to the penultimate cysteines of the mu (μ) heavy chains. The five-unit IgM molecule has a molecular weight of 970,000 kDal and has 10 antigen-combining sites that bind antigen with high avidity. In serum, the normal range for IgM is 85 to 350 mg/100 mL, and the half-life is 5 to 6 days.
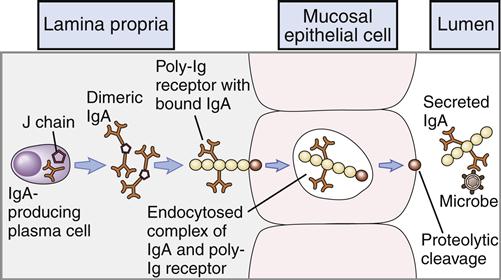
Secretory IgM (sIgM) is produced by glandular-associated B cells. During synthesis, a specialized J-chain is added to the molecule. The J chain serves two functions. Along with the Fc portion of the antibody, it is required for binding to the poly-Ig receptors (pIgRs) that mediate transcytosis through epithelial cells. In addition to its role in transportation to the exterior, the addition of the J chain causes a conformational change in the molecule, which allows the addition of an 83,000-MW fragment of the pIgR as it traverses through epithelial cells into extracellular fluids. The pIgR fragment, now called the secretory piece, protects the sIgM from enzymatic degradation.
Monomeric IgM is localized in B cell membranes and functions as a B cell antigen receptor (BCR). The membrane form has an additional 41 amino acids in a CH4 domain, and 25 hydrophobic amino acids are found in the transmembrane portion. The remaining cytoplasmic portion contains 16 polar amino acids (see Chapter 8).
The Anamnestic Response
In the lymph node, IgM-positive (IgM+) and IgG+ B cells serve as memory cells. After a second antigenic challenge, the IgG+ B cells rapidly differentiate into plasma cells that produce antigen-specific IgG antibodies. The proliferation and differentiation of memory B cells into IgG-producing plasma cells is called the anamnestic response (Figure 9-6). In isotypic switching, light chains and VH segments remain intact, but γ-chain–constant regions replace IgM-constant regions in the final antibody structure to create monomeric IgG. IgM+ memory cells function to replenish the memory cell pool. They undergo rapid proliferation and isotype switching to become IgG+ memory cells. In the anamnestic response, large concentrations of high-affinity IgG appears in serum 1 to 3 days after exposure to antigens.
Immunoglobulin G Isotype
Immunoglobulin G (IgG, or 7S) antibodies consist of two H chains and two L chains. Unlike IgM, the smaller IgG is able to penetrate extracellular and intracellular spaces. IgG binds antigens with both high affinity and low avidity. Normal adult serum levels range from 640 to 1350 mg/100 mL. In peripheral blood, the IgG antibody population has an average half-life of 23 days and is the only isotype that crosses the placenta.
Immunoglobulin G Subclasses
Subclasses IgG1, IgG2, IgG3, and IgG4 are differentiated on the basis of the size of the hinge region, position of interchain disulfide bonds, and molecular weight. IgG3 has a molecular weight of 170 kDal, whereas the other subtypes have a molecular weight of 146 kDal. IgG1 and IgG3 are usually produced in response to proteins. Carbohydrate antigens elicit the production of IgG2 and IgG4. The subclasses differ in their ability to activate complement (see Chapter 11) or bind and react to Fc receptors on phagocytic cells. Complement activation by IgG1 and IgG3 is 40 times higher than that by IgG2. The IgG4 subclass appears to inhibit complement activation.
Clinical Implications of Immunoglobulin G Subtype
Common vaccines such as those for Haemophilus, Pneumococcus, and Neisseria meningitides require a vigorous IgG2 response for host protection. In most infants, synthesis of IgG2 and IgG4 begins between 2 and 4 months of age. Some children, however, have a developmental block that prevents the production of subclasses until 2 and 6 years of age. These children may fail to produce protective antibodies after administration of the Haemophilus and meningococcal vaccines containing carbohydrate antigens.
Immunoglobulin A Isotype
Immunoglobulin A (IgA, or 11S) is a dimeric antibody found in both serum and external secretions such as tears, saliva colostrums, and intestinal secretions. The basic dimeric IgA unit consists of alpha (α) heavy chains and two light chains. Two IgA molecules are usually joined together by a J-chain. Two IgA subclasses have different bonding between heavy chains, light chains, or both and resistance to proteases secreted by microbes. Most IgA1 is found as a dimer with covalently linked heavy chains. In the IgA2 form, both the heavy and light chains are linked by ionic bonds.
Lymphoid tissues such as lymph nodes and the spleen contain a predominance of IgA1-synthesizing cells. Secretory lymphoid tissues contain a high proportion of IgA2-producing cells. In the gut, IgA2 is particularly important in the defense against Salmonella and against Vibrio cholerae. The ratio of IgA subclasses in serum or external secretions depends on the nature of the antigens that elicit their production. Thymus–dependent antigens (proteins) induce the production of IgA1 antibodies. Conversely, thymus-independent type I antigens (lipopolysaccharide endotoxins) stimulate the production of IgA2 antibodies.
Human infants are capable of synthesizing IgA at 2 to 3 weeks of age. IgA is the first line of defense against respiratory and intestinal infections.
Secretory IgA
Secretory IgA (11S) is the major antibody found in bodily secretions. It is produced by a small population of plasma cells that secrete a dimeric form of serum IgA containing a J chain. The transport of IgA from blood to external fluids is identical to that described for secretory IgM. IgA binds to a polymeric Ig Fc receptor (pIgR) on the internal surface of epithelial cells. Endocytosis creates intracellular vesicles containing IgA–pIgR complexes. The secretory piece, which is a cleavage fragment of pIgR, is added to the IgA in the intracellular vesicles. The secretory piece coils around IgA and prevents its digestion in the acidic external fluids. During exocytosis, a fragment of the secretory piece is removed, and free sIgA is secreted into external fluids. In external fluids, sIgA is dimeric with a J chain and the secretory piece. The transport process for IgA is shown in Figure 9-7.
Clinical Importance of Secretory IgA
Lymphocytes producing sIgA are localized in the mucosal associated lymphoid tissue (MALT). Because the MALT is interconnected, stimulation of one mucosal surface ultimately provides protection to all mucosal surfaces. The mechanism for providing protection to all mucosal surfaces is simple. Antigen-stimulated MALT lymphocytes enter the lymph and migrate to a regional lymph node. Antigen-specific IgA-producing B cells from the lymph node enter the bloodstream and localize in all mucosal lymphoid tissues, thereby providing widely distributed protection. Vaccine makers make use of this interconnection for their purposes. For example, intranasal administration of attenuated influenza vaccine stimulates the MALT and provides protection to all respiratory mucosal surfaces.
Immunoglobulin D (IgD)
Immunoglobulin D (IgD) is composed of two delta (δ) heavy chains and two light chains. IgD is bound to B cells via the Fc receptor or is free in serum. Because it has a half-life of 2 to 3 days, the plasma concentration is less than 1% of the total immunoglobulin in serum.
Emerging evidence suggests that IgD-producing B cells are auto-reactive lymphocytes that have escaped clonal deletion. IgD-expressing B cells may actually represent a separate lineage of B cells called B-1 lymphocytes. Autoantibodies produced by these lymphocytes react with epithelial tissue, red blood cell membranes, cellular receptors and, single-stranded or double-stranded deoxyribonucleic acid (DNA) and cause autoimmune disease. Autoantibodies produced by B-1 lymphocytes mediate disorders such as systemic lupus erythematosus (SLE), myasthenia gravis, autoimmune hemolytic anemia, and idiopathic cytopenia purpura.
Immunoglobulin E
Immunoglobulin E (IgE) is the major mediator of asthma, urticaria, and rhinitis, which are classified as immediate allergic reactions. IgE is similar to IgG in structure, but IgE has two unique features: (1) The epsilon (ε) heavy chain has a high (12%) carbohydrate content and has an additional constant region (CH4). The unique CH4 region restricts IgE binding to high-affinity receptors (Fcε-RI) on basophiles and mast cells, which contain preformed granules of heparin and histamine.
Antigen-induced cross-linkage of receptor-bound IgE initiates a process that culminates in the release of histamine and heparin, which increase vascular permeability and promote contraction of smooth muscle. In addition to a role in allergic reactions, IgE plays a critical role in the immune response to parasites such as Schistosoma mansoni and Trichinella spirillum. Histamine induces muscular contractions in the intestine, which aid in the expulsion of parasites.
Primary Immunodeficiencies
Transient Hypogammaglobulinemia of Infancy
In most infants, transient hypogammaglobulinemia of infancy (THI) is short lived and related to the catabolism rate of passively acquired maternal antibodies that provide protective immunity in the infant. Rapid catabolism of maternal antibodies may occur before the infant can synthesize his or her own antibodies. The lack of antibodies creates a “physiologic antibody trough” that puts the infant at risk for infection. The trough is more pronounced in premature babies born between 26 and 32 weeks of gestation. At 3 to 6 months, the infant begins to synthesize antibodies.
Some infants also have a developmental block that prevents both IgG and IgA synthesis until reaching the age of 6 years. The pathophysiology of the developmental block is not fully delineated, but it may be caused by the aberrant synthesis of cytokines. Infants with THI have increased synthesis of tumor necrosis factor alpha (TNF-α), TNF-β, and interleukin 10 (IL-10). Increased TNF suppresses the IgG and IgA responses. IL-10 induces isotypic switching from the IgG family to IgD. Infants with THI cannot produce antibodies to carbohydrate antigens and are at risk for developing infections with Streptococcus pneumoniae and Haemophilus influenzae type B.
Common Variable Immunodeficiency
Common variable immunodeficiency (CVID) is a primary immunodeficiency. The prevalence of CVID in the United States is approximately 1 per 50,000 live births. This immunodeficiency is characterized by a paucity of B cells that are capable of producing antibodies. Molecular defects in surface interactions and signaling pathways are common in CVID pathways. Patients have reduced numbers of cell surface CD40, which is critical to B cell proliferation. Defective protein kinase C activation and tyrosine phosphorylation also are found in the BCR signaling pathway. Individuals with CVID have recurrent infections with Haemophilus influenzae, Streptococcus pneumoniae, Moraxella catarrhalis, and Staphylococcus aureus. Some patients also may have infections by uncommon pathogens such as Pneumocystis carinii, Giardia lamblia, and Mycoplasma pneumoniae.
Treatment of IgG Immunodeficiencies
For most primary immunoglobulin immunodeficiencies, the cycle of recurrent infections can be interrupted by immunoglobulin replacement therapy. However, patients with IgG subclass immunodeficiency are usually not treated with immunoglobulin replacement therapy unless they fail to respond to both protein and polysaccharide antigens. Aggressive antibiotic therapy is indicated for preventing S. pneumoniae and H. influenzae infections. Patients with immunoglobulin deficiencies also have a high frequency of G. lamblia infections, which require a course of metronidazole to reduce diarrhea.
Selective IgA Deficiency
IgA deficiency is a common immunodeficiency with an estimated frequency between 1 per 200 and 1 per 1000 live births. In children, recurrent infections are not normally associated with IgA deficiency, which can be attributed to a compensatory mechanism involving secretory IgM. Many adults with IgA deficiency, however, have recurrent otitis media, sinusitis, bronchitis, and gastrointestinal (GI) infections. Between 10% and 40% of adults also have anti-IgA antibodies and have severe allergic or immune complex reactions following administration of immunoglobulins during replacement therapy.
Defects in IgA production may be the result of an intrinsic B cell defect; inadequate or defective CD4+ helper T cells; excessive IgA suppressor cells; and the suppressive effects of maternal IgA. The presence of anti-IgA antibodies in patients also suggests that a break occurs in peripheral tolerance.
Treatment of IgA Deficiency
No treatment for IgA deficiency is currently available. Antibiotic treatment of sinopulmonary and GI infections are indicated. To boost immunity, patients can be immunized with the pneumococcal polysaccharide vaccine. However, not all patients are capable of mounting an immune response to carbohydrate antigens.
Hyperimmunoglobulinemia E (Job Syndrome)
Job syndrome consists of a constellation of eczematous dermatitis, recurrent skin boils, skin abscesses, and cystic lung disease caused by S. aureus. Patients also have coarse facial features as well as abnormalities in the skeleton, connective tissue, and dentition.
Several immunologic defects are associated with Job syndrome. A reversal of the Th1:Th2 ratio in peripheral blood is seen. Th2 cells and IL-4 are elevated, which results in isotypic switching to IgE production. The low number of Th1 with defective production of cytokines and adhesion factors impairs inflammatory responses.
Treatment of Job Syndrome
Each clinical problem is usually treated separately. Topical steroids and tacrolimus are used to control skin eruptions. Drug coverage for staphylococcal infections is dependent on whether the Staphylococccus is resistant or sensitive to common cell wall inhibitors.
Plasma Cell Dyscrasias
Multiple Myeloma
Multiple myeloma is a neoplasm within the marrow of the axial skeletal system. It is characterized by the uncontrolled proliferation of a single plasma cell clone. In bone marrow, proliferation of plasma cells causes soft tissue masses (plasmacytomas). Because the plasmacytoma is derived from a single plasma cell clone, antibodies have the same isotype, antigen specificity, and affinity constant. These antibodies are called monoclonal antibodies because they are produced by a single B cell clone and have striking homogeneity. Some free antibody light chains are found in serum from patients with multiple myeloma. Light chains excreted into urine are termed Bence Jones proteins, thus named after the British physician who first described the proteins.
Bone destruction is also associated with multiple myeloma. High levels of IL-6 produced by a plasmacytoma cause the release of osteoclast-stimulating factor (OSF). Activated osteoclasts destroy the mineralized bone matrix and release calcium into blood.
Bone marrow destruction often results in compression fractures of the spine and weight-bearing bones. Increased calcium levels in the blood also damage the kidneys. Kidney failure is the leading cause of death in patients with multiple myeloma.
Treatment of Multiple Myeloma
Treatment of multiple myeloma is complicated. Several combinations of chemotherapeutics have been used to reduce the tumor mass. The most commonly used regimen is melphalan and prednisone. However, quicker therapeutic results have been reported with a combination of vincristine, Adriamycin, and dexamethasone (VAD) therapy. Bisphosphonates are also administered to increase bone healing and prevent hypocalcemia.
Waldenström’s Macroglobulinemia
Waldenström’s macroglobulinemia is a rare non-Hodgkin’s lymphoma, which causes an overproduction of monoclonal IgM. IgM levels above 40 g/L increase the viscosity of serum. Hyperviscosity causes headaches, confusion, dizziness, and deafness.
Treatment of Waldenström’s Macroglobulinemia
Plasmapheresis is used to reduce the levels of serum IgM and restore normal viscosity. Maintenance plasmapheresis may be required to keep the patient asymptomatic. Chemotherapy must also be used to control the proliferation of IgM-secreting cells.
Summary