Pseudomonas, Burkholderia, and Similar Organisms
1. Describe the normal sources (habitat) for Pseudomonas aeruginosa, Burkholderia cepacia, Burkholderia pseudomallei, and Burkholderia mallei, including the routes of transmission.
2. Identify the factors that contribute to the pathogenicity of P. aeruginosa and explain the physiologic mechanism for each.
3. List the various disease states associated with P. aeruginosa and Burkholderia spp.
4. Compare and contrast the Gram stain appearance of the gram-negative bacilli discussed in this chapter.
5. List the appropriate scheme for identifying P. aeruginosa.
6. Describe the media and chemical principle of each used, including differential and selective agars that aid the cultivation of Pseudomonas, Brevundimonas, and Ralstonia spp.
7. Describe the potential therapies for B. cepacia and B. pseudomallei and the concerns about optimal therapy.
8. Describe and identify the patterns of antibiotic resistance in P. aeruginosa.
General Characteristics
At one time, most of the species belonging to the genera Brevundimonas, Burkholderia, Ralstonia, and Acidovorax were members of the genus Pseudomonas. Organisms in these genera have many similar characteristics. They are aerobic, non–spore-forming, straight, slender, gram-negative bacilli with cells that range from 1 to 5 µm long and 0.5 to 1 µm wide. All species except B. mallei are motile, having one or several polar flagella. Members of these genera use a variety of carbohydrate, alcohol, and amino acid substrates as carbon and energy sources. Although they are able to survive and possibly grow at relatively low temperatures (i.e., as low as 4°C), the optimum temperature range for growth of most species is 30° to 37°C; that is, they are mesophilic. Burkholderia gladioli, Pseudomonas luteola, and Pseudomonas oryzihabitans are oxidase negative and are discussed in Chapter 21. Pseudomonas alcaligenes, Pseudomonas pseudoalcaligenes, Ralstonia paucula, Ralstonia gilardii, Comamonas spp. (including the former Pseudomonas testosteroni), and Delftia acidovorans (formerly Pseudomonas acidovorans) are not able to utilize glucose and are discussed in Chapter 25. Acidovorax facilis is MacConkey negative. Pseudomonas spp. are catalase positive. The organisms in this chapter are all oxidase-positive, grow on MacConkey agar, and oxidize glucose.
Epidemiology
Burkholderia spp. and Ralstonia Pickettii
Burkholderia spp. and Ralstonia pickettii are inhabitants of the environment and are not considered part of the normal human flora (Table 22-1). As such, their transmission usually involves human contact with heavily contaminated medical devices or substances encountered in the hospital setting.
TABLE 22-1
| Species | Habitat (Reservoir) | Mode of Transmission |
| Burkholderia cepacia | Environmental (soil, water, plants); survives well in hospital environment; not part of normal human flora; may colonize respiratory tract of patients with cystic fibrosis | Exposure of medical devices and solutions contaminated from the environment; person-to-person transmission also documented |
| B. pseudomallei | Environmental (soil, streams, surface water, such as rice paddies); limited to tropical and subtropical areas, notably Southeast Asia; not part of human flora | Inhalation or direct inoculation from environment through disrupted epithelial or mucosal surfaces |
| B. mallei | Causative agent of glanders in horses, mules, and donkeys; not part of human flora | Transmission to humans is extremely rare; associated with close animal contact and introduced through mucous membranes or broken skin. |
| Ralstonia pickettii | Environmental (multiple sources); found in variety of clinical specimens; not part of human flora | Mode of transmission is not known; likely involves exposure to contaminated medical devices and solutions |
| Pseudomonas aeruginosa | Environmental (soil, water, plants); survives well in domestic environments (e.g., hot tubs, whirlpools, contact lens solutions) and hospital environments (e.g., sinks, showers, respiratory equipment); rarely part of normal flora of healthy humans | Ingestion of contaminated food or water; exposure to contaminated medical devices and solutions; introduction by penetrating wounds; person-to-person transmission is assumed to occur |
| P. alcaligenes, P. pseudoalcaligenes, Pseudomonas sp. CDC group 1, “P. denitrificans,” Pseudomonas-like group 2, and CDC group Ic | Environmental; not part of normal human flora | Uncertain. Rarely encountered in clinical specimens |
| P. fluorescens, P. putida, P. stutzeri, (including Vb-3), P. luteola, and P. mendocina | Environmental (soil and water); not part of normal human flora | Exposure to contaminated medical devices and solutions |
| Brevundimonas vesicularis and B. diminuta | Environmental; not part of normal human flora | Uncertain. Rarely encountered in clinical specimens |
| Acidovorax spp. | Environmental, soil; not part of human flora | Unknown. Rarely found in humans |
Pseudomonas spp. and Brevundimonas spp.
The genera Pseudomonas and Brevundimonas comprise several environmental species that rarely inhabit human skin or mucosal surfaces. In the clinical setting, P. aeruginosa is the most commonly encountered gram-negative species that is not a member of the family Enterobacteriaceae and is an uncommon member of the normal human flora. The organism survives in various environments in nature and in homes and hospitals (see Table 22-1). Brevundimonas spp. are environmental and are encountered primarily in nature in water, soil, and on plants, including fruits and vegetables. Because of the ubiquitous nature of P. aeruginosa and Brevundimonas spp., the transmission of to humans can occur in a variety of ways.
P. fluorescens, P. putida, and P. stutzeri are environmental inhabitants, but they are much less commonly found in clinical specimens than is P. aeruginosa. The other pseudomonads and Brevundimonas spp. listed in Table 22-1 are also environmental organisms. Because they are rarely encountered in patient specimens, the mode of transmission to humans remains uncertain.
Pathogenesis and Spectrum of Disease
Burkholderia spp. and Ralstonia Pickettii
Because Burkholderia spp. and R. pickettii are uncommon causes of infection in humans, very little is known about what, if any, virulence factors they exhibit. Except for B. pseudomallei, the species listed in Table 22-2 generally are nonpathogenic for healthy human hosts.
TABLE 22-2
Pathogenesis and Spectrum of Disease
| Species | Virulence Factors | Spectrum of Disease and Infections |
| Burkholderia cepacia | Unknown. Binding of mucin from patients with cystic fibrosis may be involved. Intrinsic resistance to multiple antibiotics complicates therapy and may promote organism survival in hospital | Nonpathogenic to healthy human hosts; able to colonize and cause life-threatening infections in patients with cystic fibrosis or chronic granulomatous disease; other patients may suffer nonfatal infections of the urinary tract, respiratory tract, and other sterile body sites |
| B. pseudomallei | Unknown. Bacilli can survive within phagocytes | Wide spectrum from asymptomatic infection to melioidosis, of which there are several forms, including infections of the skin and respiratory tract, multisystem abscess formation, and bacteremia with septic shock |
| B. mallei | Unknown for human infections | Human disease is extremely rare. Infections range from localized acute or chronic suppurative infections of skin at site of inoculation to acute pulmonary infections and septicemia |
| Ralstonia pickettii | Unknown | Rarely encountered as cause of disease; nonpathogenic to healthy human host, but may be isolated from a variety of clinical specimens, including blood, sputum, and urine; when encountered environmental contamination should be suspected |
| Pseudomonas aeruginosa | Exotoxin A, endotoxins, proteolytic enzymes, alginate, and pili; intrinsic resistance to many antimicrobial agents | Opportunistic pathogen that can cause community- or hospital-acquired infections Community-acquired infections: skin (folliculitis); external ear canal (otitis externa); eye, following trauma; bone (osteomyelitis), following trauma; heart (endocarditis) in IV drug abusers; and respiratory tract (patients with cystic fibrosis) Hospital acquired infections: respiratory tract, urinary tract, wounds, bloodstream (bacteremia), and central nervous system Key pathogen that infects lungs of cystic fibrosis patients |
| P. fluorescens, P. putida, and P. stutzeri (includes Vb-3) | Unknown. Infection usually requires patient with underlying disease to be exposed to contaminated medical devices or solutions | Uncommon cause of infection; have been associated with bacteremia, urinary tract infections, wound infections, and respiratory tract infections; when found in clinical specimen, significance should always be questioned |
| P. mendocina, P. alcaligenes, P. pseudoalcaligenes, Pseudomonas sp. CDC group 1, “P. denitrificans,” Pseudomonas-like group 2, and CDC group Ic | Unknown | Not typically known to cause human infections. P. mendocina has been isolated from a patient with endocarditis (R) |
| Brevundimonas vesicularis and B. diminuta | Unknown | Rarely associated with human infections. B. vesicularis is rare cause and of bacteremia in patients suffering underlying disease |
| Acidovorax spp. | Unknown | Rarely isolated from clinical specimens. Not implicated in human infections |
The remaining species listed in Table 22-2 are rarely encountered in human disease, and their clinical significance should be questioned when they are found in clinical specimens.
Pseudomonas spp. and Brevundimonas spp.
P. aeruginosa also contains several genes involved in quorum sensing, a mechanism for detecting bacterial products in the immediate environment. When the growth of the organism or neighboring bacteria reaches a critical mass, the concentration of these “inducing” molecules reaches a level that activates transcription of virulence factors, including genes related to metabolic processes, enzyme production, and the formation of biofilm. Although many in vitro studies have examined biofilm formation, no clear evidence exists that demonstrates a clear role for biofilm in the organism’s pathogenesis. Although biofilm studies have been examined in the laboratory, it is evident that P. aeruginosa does not form the same type of biofilm in vivo as is seen on artificial surfaces. Biofilm production related to the overproduction of alginate and the mucoid phenotype isolated from patients with cystic fibrosis is associated with serious infections. P. aeruginosa forms microcolonies in tissue that are associated with quorum-sensing, biofilm-producing strains, which indicates that the quorum sensing is also linked to the formation of microcolonies. These microcolonies contain DNA, mucus, actin, and other products from dying bacterial and host cells. Additionally, P. aeruginosa can survive harsh environmental conditions and displays intrinsic resistance to a wide variety of antimicrobial agents, two factors that facilitate the organism’s ability to survive in the hospital setting (see Table 22-2).
Even with the variety of potential virulence factors discussed, P. aeruginosa remains an opportunistic pathogen that requires compromised host defenses to establish infection. In normal, healthy hosts, infection is usually associated with events that disrupt or bypass protection provided by the epidermis (e.g., burns, puncture wounds, use of contaminated needles by intravenous drug abusers, eye trauma with contaminated contact lenses). The result is infections of the skin, bone, heart, or eye (see Table 22-2).
No known virulence factors have been associated with P. fluorescens, P. putida, or P. stutzeri. When infections caused by these organisms occur, they usually involve a compromised patient exposed to contaminated medical materials. Such exposure has been known to result in infections of the respiratory and urinary tracts, wounds, and bacteremia (see Table 22-2). However, because of their low virulence, whenever these species are encountered in clinical specimens, their significance should be highly suspect. Similar caution should be applied whenever the other Pseudomonas spp. or Brevundimonas spp. listed in Table 22-2 are encountered.
Laboratory Diagnosis
Specimen Collection and Transport
No special considerations are required for specimen collection and transport of organisms discussed in this chapter. Refer to Table 5-1 for general information on specimen collection and transport.
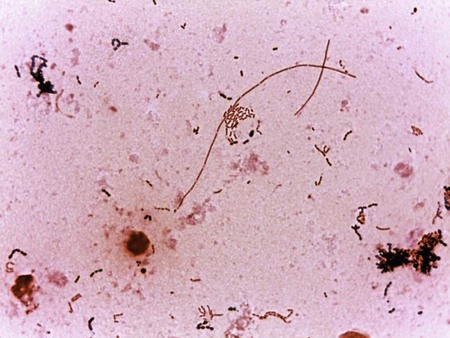
Cultivation
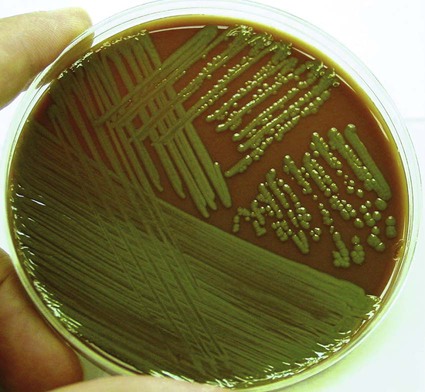
Media of Choice
Pseudomonas spp., Brevundimonas spp., Burkholderia spp., R. pickettii, and CDC group Ic grow well on routine laboratory media, such as 5% sheep blood agar and chocolate agar (Figure 22-1). Except for B. vesicularis, all usually grow on MacConkey agar. All four genera also grow well in broth-blood culture systems and common nutrient broths, such as thioglycollate and brain-heart infusion. Specific selective media, such as Pseudomonas cepacia (PC) agar or oxidative–fermentative base–polymyxin B–bacitracin–lactose (OFPBL) agar may be used to isolate B. cepacia from the respiratory secretions of patients with cystic fibrosis (Table 22-3). PC agar contains crystal violet, bile salts, polymyxin B, and ticarcillin to inhibit gram-positive and rapid-growing, gram-negative organisms. Inorganic and organic components, including pyruvate and phenol red, also are added. B. cepacia breaks down the pyruvate, creating an alkaline pH and resulting in a color change of the pH indicator (phenol red) from yellow to pink. OFPBL incorporates bacitracin as an added selective agent and uses lactose fermentation to differentiate isolates. B. cepacia ferments lactose and appears yellow, whereas nonfermenters appear green. Ashdown medium is used to isolate B. pseudomallei when an infection caused by this species is suspected. The medium contains crystal violet and gentamicin as selective agents to suppress the growth of contaminating organisms. Neutral red is incorporated into the medium and is taken up by the organism, making it distinguishable from other bacteria.
TABLE 22-3
| Organism | Medium | Appearance |
| Acidovorax delafieldii | BA | No distinctive appearance |
| Mac | NLF | |
| Acidovorax facilis | BA | No distinctive appearance |
| Mac | Unable to grow | |
| A. temperans | BA | No distinctive appearance |
| Mac | NLF | |
| Brevundimonas diminuta | BA | Chalk white |
| Mac | NLF | |
| B. vesicularis | BA | Orange pigment |
| Mac | NLF, but only 66% grow | |
| Burkholderia cepacia complex | BA | Smooth and slightly raised; dirtlike odor |
| Mac | NLF; colonies become dark pink to red due to oxidation of lactose after 4-7 days | |
| PC or OFPBL | Smooth | |
| B. pseudomallei | BA | Smooth and mucoid to dry and wrinkled (may resemble P. stutzeri) |
| Mac | ||
| Ashdown | NLF | |
| Dry, wrinkled, violet-purple | ||
| B. mallei | BA | No distinctive appearance |
| Mac | NLF | |
| Pandoraea spp. | BA | No distinctive appearance |
| Mac | NLF | |
| Pseudomonas aeruginosa | BA | Spreading and flat, serrated edges; confluent growth; often shows metallic sheen; bluish green, red, or brown pigmentation; colonies often beta–hemolytic; grapelike or corn tortilla–like odor; mucoid colonies commonly seen in patients with cystic fibrosis |
| Mac | NFL | |
| P. fluorescens | BA | No distinctive appearance |
| Mac | NLF | |
| P. mendocina | BA | Smooth, nonwrinkled, flat, brownish–yellow pigment |
| Mac | NLF | |
| P. monteilii | BA | No distinctive appearance |
| Mac | NLF | |
| P. mosselii | BA | No distinctive appearance |
| Mac | NLF | |
| No acid production from xylose | ||
| P. putida | BA | No distinctive appearance |
| Mac | NLF | |
| P. stutzeri and CDC group Vb–3 | BA | Dry, wrinkled, adherent, buff to brown |
| Mac | NLF | |
| P. veronii | BA | No distinctive appearance |
| Mac | NLF | |
| Pseudomonas–like group 2 | BAP | No distinctive appearance but colonies tend to stick to agar |
| Mac | ||
| NLF | ||
| CDC group Ic | BAP | No distinctive appearance |
| Mac | NLF | |
| Ralstonia mannitolilytica | BAP | No distinctive appearance |
| Mac | NLF | |
| R. pickettii | BAP | No distinctive appearance but may take 72 hr to produce visible colonies |
| Mac | NLF |
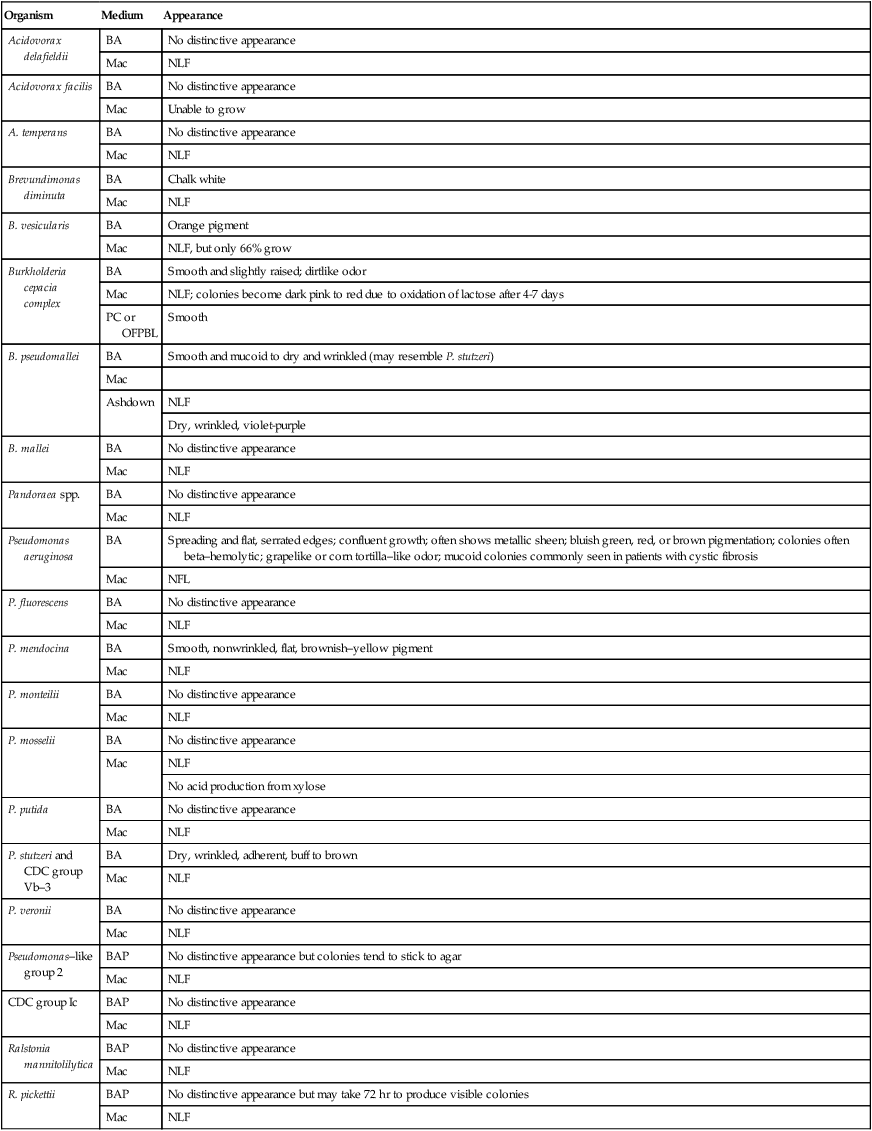
BAP, 5% sheep blood agar; Mac, MacConkey agar; NLF, non–lactose-fermenter; OFPBL, oxidative–fermentative base–polymyxin B–bacitracin–lactose; PC, Pseudomonas cepacia agar.
Approach to Identification
Table 22-4 provides the key phenotypic characteristics for identifying the species discussed in this chapter. These tests provide useful information for presumptive organism identification, but definitive identification often requires the use of a more extensive battery of tests performed by reference laboratories.
TABLE 22-4
Biochemical and Physiologic Characteristics
| Organisms | Growth at 42°C | Nitrate Reduction | Gas from Nitrate | Gelatin Liquefied | Arginine Dihydrolase | Lysine Decarboxylase | Urea Hydrolysis | Oxidizes Glucose | Oxidizes Lactose | Oxidizes Mannitol | Oxidizes Xylose |
| Acidovorax delafieldii | v | + | − | − | + | − | + | + | − | v | v |
| Acidovorax facilis | − | + | − | + | + | − | + | + | − | + | + |
| Acidovorax temperans | + | + | − | − | − | − | v | + | − | v | − |
| Brevundimonas diminuta | v | − | − | v | − | − | − | v | − | − | − |
| B. vesicularis | v | − | − | v | − | − | − | v | − | − | v |
| Burkholderia cepacia complex | v | v | − | v | + | v | v | + | v | + | v |
| B. pseudomallei | + | + | + | v | + | − | v | + | + | + | + |
| B. mallei | − | + | − | − | + | − | v | + | v | − | v |
| Pandoraea spp. | v | v | − | − | − | − | v | +w | − | − | − |
| Pseudomonas aeruginosa | + | + | + | v | + | − | v | + | − | v | + |
| P. fluorescens | − | − | − | + | + | − | v | + | v | v | + |
| P. mendocina | + | + | + | − | + | − | v | + | − | − | + |
| P. monteilii | − | − | − | − | + | − | v | + | − | − | − |
| P. mosselii | − | − | − | + | + | − | ND | + | − | v | − |
| P. putida | − | − | − | − | + | − | v | + | v | v | + |
| P. stutzeri | v | + | + | − | − | − | v | + | − | + | + |
| Pseudomonas veronii | − | + | + | v | + | ND | v | + | ND | + | + |
| Pseudomonas–like group 2 | v | v | − | − | v | − | + | + | + | + | + |
| CDC group Ic | + | + | − | − | + | − | v | + | − | − | − |
| Ralstonia insidiosa | + | + | ND | ND | N | − | v | − | v | ND | ND |
| Ralstonia mannitolilytica | + | − | − | v | − | − | + | + | + | + | + |
| R. pickettii | v | + | v | v | − | − | + | + | v | − | + |
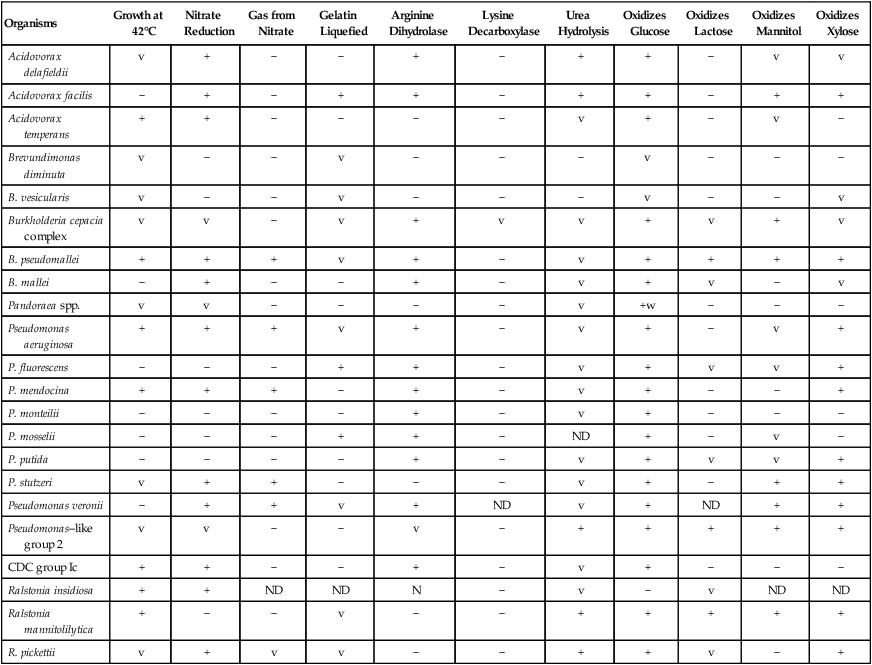
ND, No data; v, variable; +, >90% of strains are positive; −, >90% of strains are negative; w, weak.
*Arginine-positive strains of P. stutzeri formerly classified as CDC group Vb-3.
Comments Regarding Specific Organisms
• Triple sugar iron slant with an alkaline/no change (K/NC) reaction
• Production of bright bluish (pyocyanin) green (pyoverdin), red (pyorubin), or brown (pyomelanin) diffusible pigment on Mueller-Hinton agar or trypticase soy agar (Figures 22-2 and 22-3)
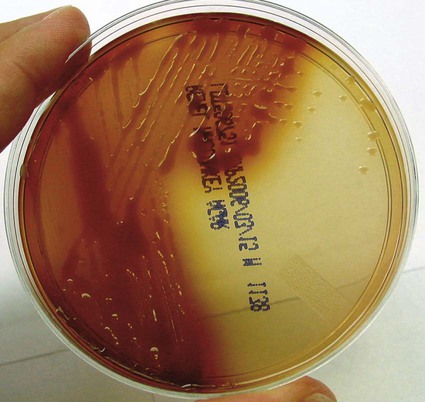
The presumptive identification of other species in this chapter is fairly straightforward using the key characteristics given in Table 22-4. However, a few notable exceptions exist. First, when B. cepacia complex is identified by a commercial system in a patient with cystic fibrosis, species confirmation should be completed by a combination of phenotypic and genotypic methods. This is also true if a rapid system identifies an organism as B. gladioli or R. pickettii. The B. cepacia complex has 10 genomovars, and appropriate speciation is crucial.
Antimicrobial Susceptibility Testing and Therapy
Many of these organisms grow on the media and under the conditions recommended for testing of the more commonly encountered bacteria (see Chapter 12 for more information about validated testing methods); however, the ability to grow under test conditions does not guarantee reliable detection of important antimicrobial resistance. Therefore, even though testing can provide an answer, it poses a substantial risk of erroneous interpretations. Validated susceptibility testing methods are available for a limited number of antibiotics.
Among Pseudomonas spp. and Brevundimonas spp., P. aeruginosa is the only species for which valid in vitro susceptibility testing methods exist and for which extensive therapeutic evidence exists (see Table 22-5; also see Chapter 12 for a discussion of available testing methods). Therapy usually involves the use of a beta-lactam developed for antipseudomonal activity and an aminoglycoside. The particular therapy used depends on several clinical factors and on the laboratory antimicrobial resistance profile for the P. aeruginosa isolate. P. aeruginosa isolated from patients with cystic fibrosis may require extended incubation for up to 24 hours before obtaining a reliable susceptibility pattern. In addition, the organism may develop resistance during prolonged therapy with any antimicrobial agent within 3 to 4 days requiring repeat susceptibility testing.
TABLE 22-5
Antimicrobial Therapy and Susceptibility Testing
| Species | Therapeutic Options | Potential Resistance to Therapeutic Options | Validated Testing Methods* | Comments |
| Burkholderia cepacia | Potentially active agents include piperacillin, ceftazidime imipenem, ciprofloxacin, chloramphenicol, and trimethoprim/sulfamethoxazole | Yes | Disk diffusion, broth dilution, and E-tests | Antimicrobial therapy rarely eradicates organism. Development of resistance during therapy may warrant additional susceptibility testing. |
| B. pseudomallei | Potentially active agents include ceftazidime, piperacillin/tazobactam, ticarcillin/clavulanate, amoxicillin/clavulanate, imipenem, trimethoprim/sulfamethoxazole, and chloramphenicol | Yes | Disk diffusion, broth dilution, agar dilution, and E-tests | Disk diffusion testing for TMP-SMX is unreliable |
| B. mallei | No definitive guidelines Potentially active agents. may include those listed for B. pseudomallei | Yes | Disk diffusion, broth dilution, agar dilution, and E-tests | Relapses may occur following therapy |
| Ralstonia pickettii | No definitive guidelines. Potentially active agents include those listed for B. cepacia | Yes | Not available | Rarely involved in human infections, so reliable therapeutic data are limited |
| Pseudomonas aeruginosa | An antipseudomonal beta-lactam (listed below) with or without an aminoglycoside; certain quinolones may also be used. Specific agents include piperacillin/tazobactam, ceftazidime, cefepime, aztreonam, imipenem, meropenem, gentamicin, tobramycin, amikacin, netilmicin, ciprofloxacin, and levofloxacin | Yes | Disk diffusion, broth dilution, agar dilution, and commercial systems | In vitro susceptibility testing results important for guiding therapy |
| P. fluorescens, P. putida, P. stutzeri (includes Vb-3), P. mendocina, P. alcaligenes, P. pseudoalcaligenes, Pseudomonas sp. CDC group 1, “P. denitrificans,” Pseudomonas-like group 2, and CDC group Ic | Because rarely implicated in human infections, there are no definitive guidelines; agents used for P. aeruginosa may be effective for these species | Yes | Not available | Most will grow on susceptibility testing media, but standards for interpretation of results do not exist |
| Pseudomonas luteola P. oryzihabitans |
No definitive guidelines. Potentially active agents include cefotaxime, ceftriaxone, ceftazidime, imipenem, quinolones, and aminoglycosides | Yes, activity of penicillins is variable; commonly resistant to first- and second-generation cephalosporins | Not available | |
| Brevundimonas vesicularis B. diminuta |
Because rarely implicated in human infections, there are no definitive guidelines | Unknown | Not available | Rarely involved in human infection |
| Acidovorax spp. | No definitive guidelines | Unknown | Not available | No clinical experience |
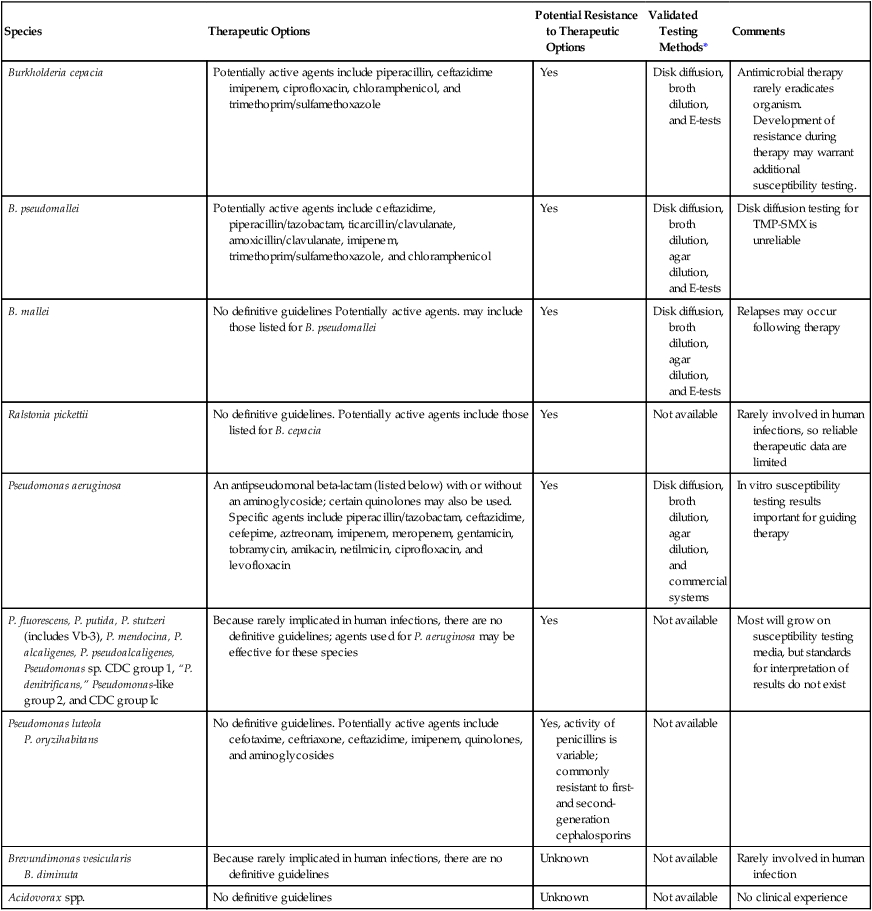
TMP-SMX – Trimethoprim-Sulfamethoxazole.
*Validated testing methods include standard methods recommended by the Clinical and Laboratory Standards Institute (CLSI) and commercial methods approved by the U.S. Food and Drug Administration (FDA).
P. aeruginosa is intrinsically resistant to various antimicrobial agents; only those with potential activity are shown in Table 22-5. However, P. aeruginosa also readily acquires resistance to the potentially active agents listed, necessitating susceptibility testing for each clinically relevant isolate.
Although antimicrobial resistance is also characteristic of the other Pseudomonas spp. and Brevundimonas spp., the fact that these organisms are rarely clinically significant and the lack of validated testing methods prohibit the provision of specific guidelines (see Table 22-5). Antimicrobial agents used for P. aeruginosa infections are often considered for use against the other species; however, before proceeding with the development of treatment strategies, the first critical step should be to establish the clinical significance of the organism.
1. Which of the following has a Gram stain morphology that resembles safety pins?
2. Which of the following does not grow on MacConkey agar?
3. Which of the following is the key pathogen that infects the lungs of patients with cystic fibrosis?
4. Valid susceptibility testing methods are available for which organism?
5. All of the following are true of Pseudomonas and Burkholderia spp. except:
6. Which factor contributes to the pathogenicity of P. aeruginosa?
7. A reliable identification scheme for P. aeruginosa shows all of the following except:
c. Production of bluish green pigment on Mueller-Hinton agar
d. Triple sugar iron slant with an alkaline/no change reaction
8. PC agar contains crystal violet, bile salts, and polymyxin B to:
a. Inhibit gram-positive organisms
b. Inhibit slow-growing gram-negative organisms
9. Which of the following is a valid testing method for P. aeruginosa?
_____ PC or OFPBL should be used for patients with cystic fibrosis.
_____ P. aeruginosa can be distinguished from other pseudomonads by growing at 25°C.
_____ B. cepacia should be suspected whenever a nonfermentative organism that decarboxylates lysine is encountered.
11. Matching: Match each term with the corresponding description.