CHAPTER 18 Posttranslational Targeting of Proteins*
Protein synthesis is largely a monopoly of cytoplasmic ribosomes that provide all of the proteins for the nucleus, cytoplasm, peroxisomes, and secretory pathway. Even mitochondria and chloroplasts import most of their proteins from cytoplasm, despite the fact that they originated as bacterial endosymbionts and have retained the capacity to synthesize a few of their proteins. Most of the original bacterial genes moved to the nucleus of the eukaryotic host.
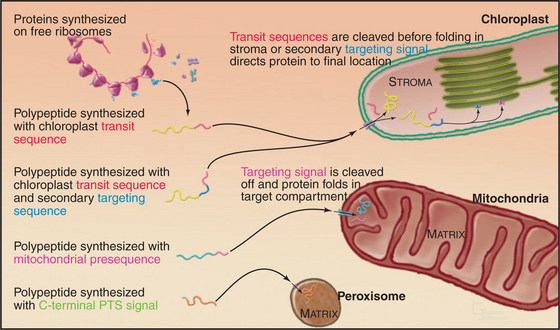
Given a common site of synthesis, accurate addressing is essential to direct proteins to their sites of action and to maintain the unique character of each cellular compartment. This is achieved by “zip codes” built into the structure of each protein (Fig. 18-1). Residues in the sequence of each protein—often, but not necessarily, contiguous amino acids—form a signal for targeting.
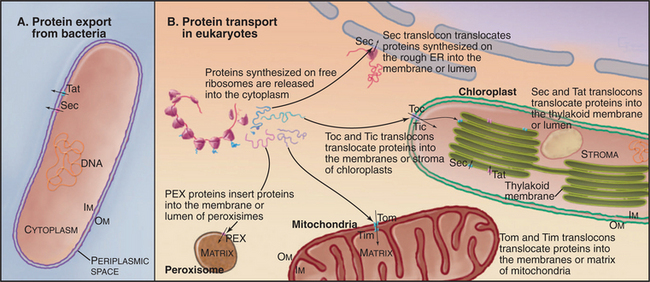
Targeting signals direct proteins to their destination by binding to organelle-specific receptors or using soluble “escort” factors as intermediaries. When necessary, proteins cross membranes via channels called translocons formed by integral membrane proteins (Fig. 18-2). Like ion channels (see Chapter 10), these protein-translocating channels are gated to prevent indiscriminate transport of cellular constituents when not occupied by a polypeptide. Polypeptides fit so tightly in these channels during translocation that ions do not leak through. Ions traverse ion channels in a microsecond, whereas polypeptides take tens of seconds to move through translocons. Protein synthesis, adenosine triphosphate (ATP) hydrolysis, or the membrane potential provides the energy to power protein translocation across membranes.
Three families of protein translocation channels are found in all three domains of life. Sec translocons direct proteins into the endoplasmic reticulum in eukaryotes and out of prokaryotes. The Tat family of pores translocate folded proteins into chloroplast thylakoids and out of prokaryotes. Membrane proteins related to Oxa1p help to insert proteins synthesized in the mitochondrial matrix and prokaryotic cytoplasm into membranes. Mitochondria (Fig. 18-4), chloroplasts (Fig. 18-6), and prokaryotes (Fig. 18-10) have additional families of protein translocation channels.
Primary targeting can occur either cotranslationally, coincident with protein synthesis, or posttranslationally, after polypeptide synthesis. Chapter 20 covers protein targeting to endoplasmic reticulum where, with a few exceptions, targeting is cotranslational. This chapter covers posttranslational targeting mechanisms that move proteins across membrane bilayers into mitochondria, chloroplasts, and peroxisomes and out of Bacteria. Eukaryotes also secrete a few proteins directly across the plasma membrane. Chapter 14 covers posttranslational movements of proteins into and out of the nucleus through a large aqueous channel in the nuclear pore.
Transport of Proteins into Mitochondria
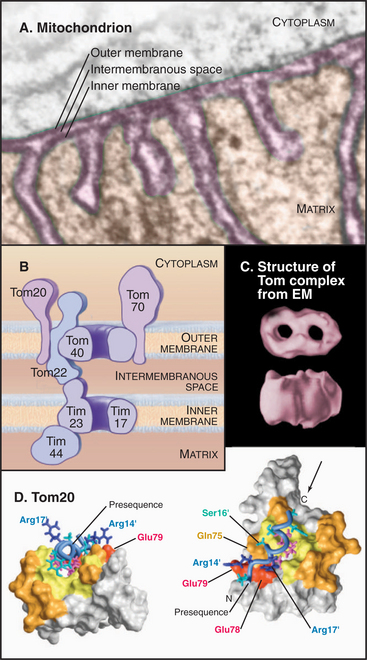
Mitochondrial outer and inner membranes define two spaces: one between the outer and inner membranes (intermembranous space) and an interior space termed the matrix (Fig. 18-3). Each membrane and space has distinct functions and protein compositions, which are covered in Chapter 19. Targeting signals and specific translocation machinery guide more than 500 imported proteins selectively to these compartments.
Genetic and biochemical experiments on fungi defined the molecular machinery for proteins to enter mitochondria, including the Tom complex (translocase of the outer mitochondrial membrane), the Sam complex (sorting and assembly machinery of the outer membrane), and two Tim complexes (translocase of the inner mitochondrial membrane). See Figures 18-4 and 18-5. Although the distinction is not absolute, one Tim complex is specialized to transport proteins into the matrix, and the other is specialized for insertion of proteins into the inner membrane. Translocation requires energy and assistance from protein chaperones both outside and inside mitochondria.
Delivery of Protein to Mitochondria
After synthesis by cytoplasmic ribosomes, most proteins destined for mitochondria bind cytosolic chaperones of the Hsp70 family (see Fig. 17-14). This interaction maintains proteins in unfolded configurations competent for import. Some imported proteins require additional factors, such as mitochondria-import stimulation factor, for targeting to the translocation machinery.
Targeting signals for proteins of the matrix are generally located at the N-termini of precursor polypeptides as contiguous sequences of 10 to 70 amino acids. These targeting motifs are called presequences, because they are usually removed by proteolytic cleavage in the mitochondrial matrix. Presequences are rich in basic, hydroxylated, and hydrophobic amino acids but share no sequences in common. The targeting sequences of many mitochondrial membrane proteins are in the middle of the polypeptide and are not cleaved after import. Cytochrome c, a component of the electron transport chain in the intermembranous space (see Fig. 19-5), also has an internal signal for import into mitochondria.
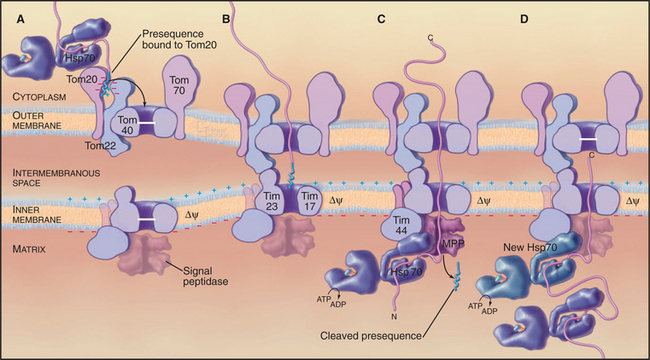
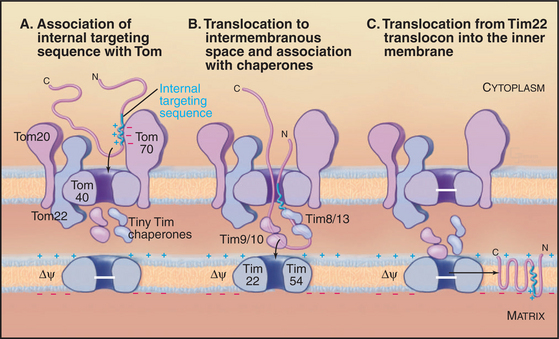
A succession of weak interactions with outer membrane receptors Tom20, Tom22, Tom5, and perhaps Tom70 guide presequences and other target signals to the outer membrane translocon. The presequence initially contacts Tom20. Eight residues of the presequence fold into an amphipathic (hydrophobic on one side, hydrophilic on the other) α-helix that binds in a shallow hydrophobic groove on Tom20. Arginines on the surface of this helix interact with acidic residues on Tom22 (Fig. 18-4D). Other parts of the presequence are thought to interact with Tom40, the translocon itself. Although these associations are weak, collectively, they distinguish mitochondrial presequences from other proteins in the cytoplasm with high fidelity.
Translocation across the Outer Membrane
Outer membrane receptors transfer the presequence to the translocon channel, which is composed mainly of Tom40 along with three small subunits. Tom40 is an integral membrane protein that is predicted to span the bilayer exclusively as β-strands. Electron microscopy of purified Tom complex revealed two pores with diameters of approximately 2 nm, which agrees with the size of the pore calculated from ion conductance measurements of purified Tom40 inserted into lipid bilayers. Two molecules of Tom40 are postulated to form a channel and the complex may contain two or three of these channels. Proteins must be largely unfolded to fit through a pore of this size. Like Sec translocons of endoplasmic reticulum (see Fig 20-6) and bacteria (Fig. 18-9), Tom channels are likely to be gated, so they close when not occupied by a translocating polypeptide. After crossing the outer membrane, some proteins remain in the intermembranous space.
Assembly of Outer Membrane Proteins
Some simple outer membrane proteins transfer laterally into the bilayer while they are in transit through Tom, while more complicated outer membrane proteins, including Tom40 itself and porins (see Fig. 7-8), require assistance. Two protein complexes of the outer membrane called Sam I and Sam II mediate folding and insertion into the membrane.
Translocation across the Inner Membrane to the Matrix
Proteins use the Tim23 translocon to cross the inner membrane into the matrix. The channel across the inner membrane is formed by the integral membrane proteins Tim23 and Tim17 (Fig. 18-4). Interactions of the N-terminal presequences of matrix proteins with Tim50 and Tim23 guide the presequence into the translocation channel. Physical interactions of Tom and Tim complexes may facilitate the transfer of matrix proteins across both membranes. The MPP peptidase (matrix processing protease) cleaves off the presequences once they enter the matrix.
Two energy sources—the electrical potential across the inner membrane and ATP hydrolysis by matrix chaperones—power polypeptide translocation across the inner membrane. The membrane potential (negative inside) pulls positively charged presequences across the membrane. Then the chaperone Hsp70 takes over and uses cycles of peptide binding and ATP hydrolysis to move the peptide into the matrix. One idea is that Hsp70 rectifies movements of the polypeptide in the pore, allowing movement forward into the matrix but not backward. Hsp70 binds when the polypeptide slides forward. After ATP hydrolysis, Hsp70 dissociates from the polypeptide and the exchange factor mGrp1 (see Fig. 17-14) rapidly recharges it with ATP, ready for another cycle of peptide binding, ATP hydrolysis, and release. This allows the polypeptide to slide forward into the matrix but not backward, so it eventually ends up as a folded protein in the matrix. Another model proposes that the energy from ATP hydrolysis is used to pull the polypeptide across the inner membrane.
Translocation into the Inner Membrane Bilayer
The integral proteins of the inner membrane lack cleaved targeting signals, depending instead on targeting information contained in the intact protein to reach their destination. One example is the most abundant protein of the inner membrane, the adenosine diphosphate (ADP)/ATP antiporter that spans the inner membrane six times (see Fig. 9-2). Its signal sequence is located in the middle of the polypeptide. A family of small “tiny Tim” chaperone proteins guide inner membrane proteins from Tom across the intermembranous space to the Tim22 translocon in the inner membrane (Fig. 18-4). This family of chaperones includes Tim8, Tim9, Tim10, Tim12, and Tim13. Complexes of Tim9/10 or Tim8/13 bind to hydrophobic segments of polypeptides during transit to the inner membrane.
Export from the Matrix
Insertion of proteins synthesized in the matrix into the inner membrane depends on an inner membrane pro-tein called Oxa1p, which forms a translocon similar to bacterial YidC and chloroplast Alb3 (see later sections). Oxa1p interacts with mitochondrial ribosomes, so it might guide hydrophobic transmembrane segments directly into the bilayer. At least one other protein complex participates in export of proteins from the matrix.
Transport of Proteins into Chloroplasts
Eukaryotes acquired chloroplasts through symbiosis with a photosynthetic cyanobacterium (see Figs. 2-8 and 19-7). Over time, most of the bacterial genes moved to the nucleus, so most chloroplast proteins are synthesized on cytoplasmic ribosomes and imported into one of three chloroplast membranes or the compartments that they surround (Fig. 19-7). Chapter 19 covers chloroplast functions. The innermost thylakoid membranes contain the photosynthetic apparatus inherited from cyanobacteria. The outer membrane likely came from the eukaryotic host, whereas the inner envelope membrane has both bacterial and eukaryotic features. Some organisms acquired their photosynthetic plastids by secondary or even tertiary rounds of endosymbiosis, when a eukaryote such as the precursor of Euglena took up a green alga (see Fig. 2-8). These secondary or tertiary plastids are bounded by one or more additional membranes and have more complicated mechanisms to import the proteins expressed from nuclear genes.
Although both chloroplasts and mitochondria arose from symbiotic Bacteria, chloroplasts evolved a distinct mechanism of protein import (Fig. 18-6). The principles are similar, but the two systems share no common proteins. The closest known relatives of any protein component of the chloroplast import machine are found in the ancestors of chloroplasts, photosynthetic cyanobacteria, in which they appear to have a role in secretion.
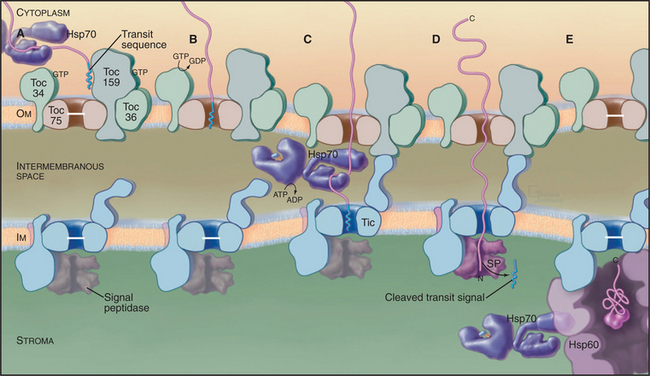
In plants, N-terminal signal sequences called transit sequences target chloroplast proteins to the import machinery in the outer envelope. When added experimentally to the N-terminus of a test protein, transit sequences suffice to guide the test protein into the stroma of chloroplasts. These N-terminal targeting se-quences are reminiscent of sequences that target proteins to endoplasmic reticulum and mitochondria, but the sequence determinants of the chloroplast transit sequences are much less well defined. They vary in length from 20 to 120 residues, and the amino acid sequences have little in common beyond a net positive charge and numerous serines and threonines.
All imported proteins use the same “general import pathway” to cross the outer and inner envelope membranes. The machinery consists of different protein complexes in each membrane called Toc (translocon at the outer envelope membrane of chloroplasts) and Tic (translocon at the inner envelope membrane of chloroplasts) (Fig. 18-6). These complexes were identified in biochemical experiments in which isolated chloroplasts imported precursor proteins synthesized in vitro. Chemical cross-linking of these imported proteins to translocon proteins identified the subunits that bind the transit sequence and contact imported polypeptides as they cross both membranes. Other subunits account for the requirements for ATP and guanosine triphosphate (GTP) hydrolysis. Both Toc and a “super complex” of Toc with Tic can be isolated for analysis of their composition. Mutations that compromise chloroplast import have also contributed to understanding the process.
As proteins emerge into the stroma, a signal peptidase cleaves off the transit peptide before the proteins fold or redistribute to their final locations. Some proteins fold with the help of Hsp70, an Hsp100 chaperone, and an Hsp60 chaperone similar to GroEL (see Fig. 17-16) and remain in the stroma. Other proteins move on to thylakoid membranes or the thylakoid lumen using at least four different pathways.
Some photosynthesis proteins insert directly into thylakoid membranes from the stroma. Others require help from proteins homologous to parts of the signal recognition particle (SRP) system used for export from bacteria (Fig. 18-10) and into the endoplasmic reticulum of eukaryotes (see Fig. 20-3). Although chloroplasts lack SRP RNA, GTPases similar to an SRP protein and the SRP receptor cooperate with a protein that is homologous to Oxa1p to mediate insertion into the thylakoid membrane.
Hydrophilic proteins destined for the thylakoid lumen retain a secondary N-terminal signal sequence after the transit sequence is cleaved in the stroma. Some move across the thylakoid membrane into the thylakoid lumen through a translocon homologous to bacterial SecYE, powered by ATP hydrolysis by a homolog of SecA (Fig. 18-9). Other proteins with tightly bound redox factors cross the thylakoid membrane while compactly folded using translocon factors similar to the bacterial Tat system (Fig. 18-2). Secondary signal sequences with two arginine residues direct these proteins to a Tat translocon and the proton gradient drives the polypeptide across the membrane. After translocation, a peptidase in the thylakoid lumen removes both types of secondary signal sequences.
Transport of Proteins into Peroxisomes
Peroxisomes are simple organelles with a single membrane limiting a lumen containing many oxidative enzymes (see Fig. 19-10). Nuclear genes encode all proteins found in the membrane and lumen of peroxisomes. Their mRNAs are translated on cytoplasmic ribosomes, and the proteins are incorporated posttranslationally into peroxisomes (Fig. 18-1).
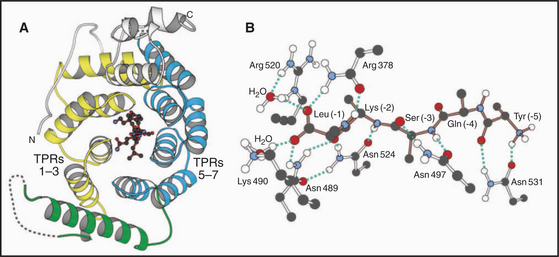
Two types of targeting signals direct proteins to the peroxisome lumen (called matrix). The type-1 peroxisomal targeting signal (PTS1) is found at the extreme C-terminus of most peroxisomal enzymes (Fig. 18-7). PTS1 is just three amino acids long, and it conforms to the consensus sequence of serine-lysine-leucine-COOH, or a conservative variant. For example, alanine or cysteine can substitute at the -3 position, arginine or histidine can function at the penultimate position, and methionine can substitute for the C-terminal leucine. PTS1 is always located at the extreme C-terminus, and amidation of the C-terminal carboxylate inactivates the signal. The type-2 peroxisomal targeting signal (PTS2) also targets proteins to the peroxisome matrix but is found on few proteins (only four are known in humans, one in yeast). PTS2 sequences are located at or near the N-terminus and have a loose consensus sequence of RLXXXXXH/QL (where X is any amino acid).
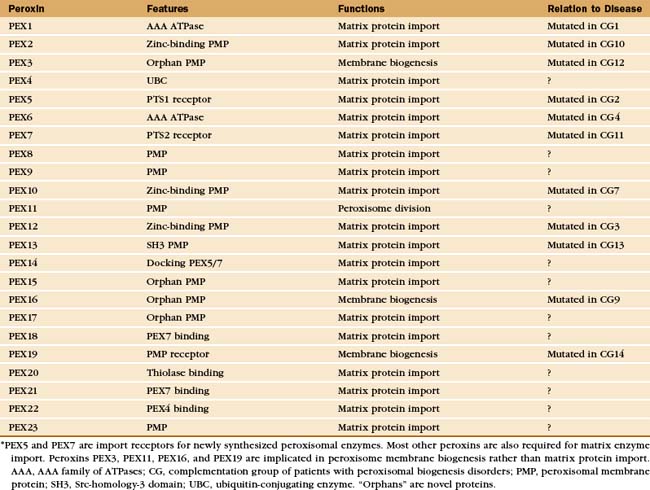
Proteins called peroxins recognize newly synthesized peroxisomal proteins and deliver them to peroxisomes for insertion into the peroxisomal membrane or translocation across the membrane into the lumen (Fig. 18-8 and Appendix 18-1). Loss of function mutations in humans and yeast revealed the genes for more than 20 peroxins that are crucial for the biogenesis and proliferation of peroxisomes. Mutations of these PEX genes in humans cause a number of devastating human diseases known as the peroxisomal biogenesis disorders (see Chapter 19).
Both PTS receptors and their cargo proteins inter-act with PEX14 and other peroxins on the peroxisomal membrane, but the mechanism that translocates proteins into the lumen is not well characterized. The translocon itself has not yet been identified, and it is not clear how large folded proteins can cross the membrane. Following translocation of a peroxisomal enzyme into the lumen, the receptors recycle back to the cytoplasm for further rounds of import.
Cells depend on a different set of peroxins, including PEX3, PEX16, and PEX19, to insert proteins into the peroxisomal membrane (Fig. 18-8). Cells that are deficient in any of these three peroxins lack peroxisomal membranes and the peroxisomal membrane proteins are degraded or mislocalized to other cellular membranes, particularly mitochondria. PEX3 is an integral protein of the peroxisomal membrane. Some PEX16 is in the cytoplasm; some is attached to the cytoplasmic side of the membrane by a farnesyl tag. PEX19 plays a dual role: As a cytoplasmic chaperone, it binds and stabilizes peroxisomal membrane proteins in cytoplasm; and as an import receptor, it recruits proteins with mPTS sequences to the peroxisomal membrane.
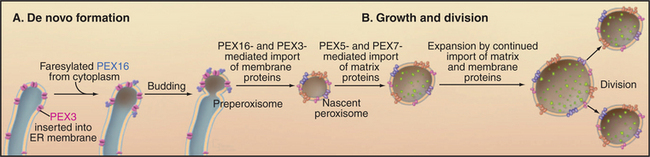
Peroxisomes may arise by either of two pathways (Fig. 18-8). Peroxisomes can form de novo by budding from the ER. PEX3 is inserted into the ER, where it recruits PEX16 and other peroxins. This specialized domain of ER then pinches off for delivery to peroxisomes or to form a nascent peroxisome de novo. By originating from the ER in this manner, peroxisomes can arise in cells that lack them without a preexisting peroxisome as template. Preexisting peroxisomes can grow by importing proteins and lipids and then divide by a process of fission dependent on the GTPase dynamin (see Fig. 22-11).
Translocation of Eukaryotic Proteins across the Plasma Membrane by ABC Transporters
Most proteins that are secreted by eukaryotic cells travel to the cell surface through the classical secretory pathway, including the endoplasmic reticulum and Golgi apparatus (see Chapters 20 and 21). But budding yeast use an ABC transporter (see Fig. 8-9) to transport their α-type mating factor directly from the cytoplasm across the plasma membrane. The α-factor is synthesized in the cytoplasm as part of a precursor, excised from the precursor by proteolytic cleavage, and then prenylated on its C-terminus before transport across the plasma membrane. This mechanism has been invoked to explain the secretion of a few mammalian proteins that lack the “signal sequences” that direct proteins to the classic ER secretory pathway. These include some cytokines, fibroblast growth factor, and some blood-clotting factors. This is a well-characterized route for secretion of some bacterial proteins (Fig. 18-10).
Targeting to the Surfaces of the Plasma Membrane
Many proteins synthesized in the cytoplasm are targeted to the cytoplasmic side of organelle and plasma membranes (see Fig. 7-9). These include peripheral membrane proteins that bind to cytoplasmic domains of integral membrane proteins or bind directly to the lipid bilayer.
Proteins attached to the external surface of plasma membranes by glycosylphosphatidylinositol anchors arrive by a different route. These proteins are synthesized on ribosomes associated with the endoplasmic reticulum and then translocated into the ER lumen anchored by a C-terminal transmembrane segment. Inside the ER the protein is cleaved from its membrane anchor and transferred enzymatically to glycosylphosphatidylinositol before transport to the cell surface (see Fig. 20-7C).
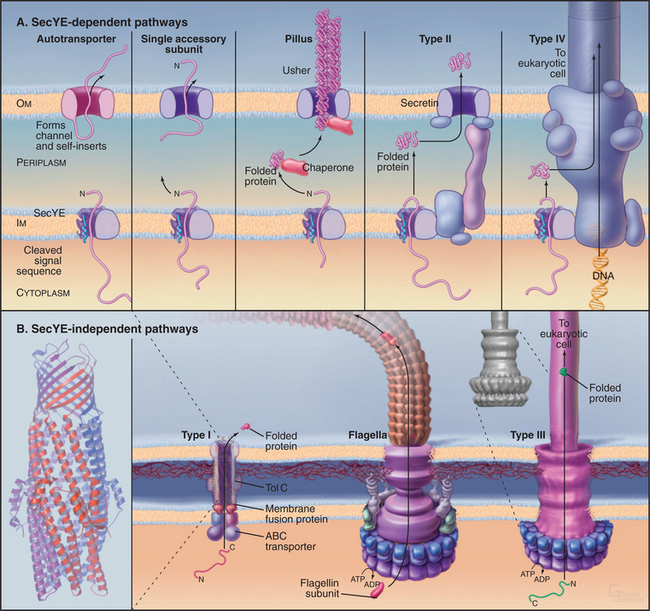
Bacterial Protein Export
Bacteria employ at least 10 distinct strategies to transport proteins from the cytoplasm across the inner membrane and beyond. Seven of these pathways use a common pore across the inner membrane called the Sec translocon. These pathways are important because some contribute to human disease. In addition, they serve as important model systems, as eukaryotes use a homologous translocon to move proteins into the bilayer or lumen of the endoplasmic reticulum (see Fig. 20-6). This section begins with a discussion of six branches of the Sec secretory pathway and finishes with three distinct pathways.
Pathways Dependent on the SecYE Translocon
Organisms in all three domains of life use Sec translocons to move proteins synthesized in the cytoplasm across membranes. Translocons in the plasma membranes of Bacteria and Archaea consist of two transmembrane proteins called SecY and SecE in Bacteria (Fig. 18-9). The translocons of the endoplasmic reticulum of eukaryotes consist of homologous protein subunits called Sec61a and g (see Fig. 20-6). The narrow pore for translocating the secreted polypeptide is located in the middle of a bundle of α-helices. Loss of function mutations of SecY or SecE compromise the secretion of most proteins by Bacteria or Archaea. Several accessory subunits assist in translocation, but they are not essential in Bacteria or present in eukaryotes.
Posttranslational Protein Translocation
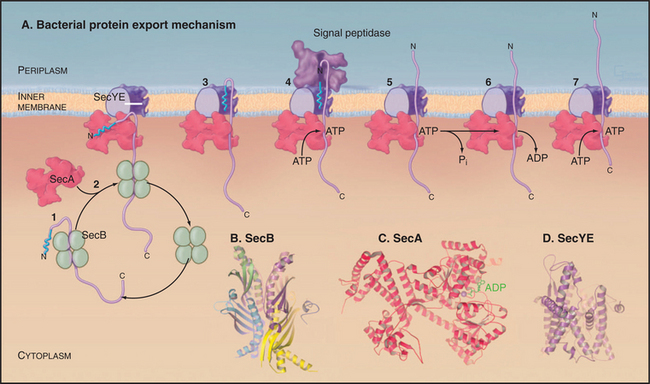
Bacteria use Sec-signal sequences to direct many proteins to the SecYE translocon for transport across the plasma membrane or for insertion into the plasma membrane. Gram positive bacteria such as Bacillus subtilis lack an outer membrane, so the proteins leave the cell after crossing the plasma membrane. In gram-negative bacteria, translocated proteins enter the periplasm, insert into the outer membrane, or leave the cell.
Proteins targeted to the Sec translocon are synthesized in the cytoplasm with an N-terminal Sec-signal sequence. These targeting sequences consist of about 25 residues beginning with methionine, followed by a few basic residues, 10 to 15 hydrophobic residues, and a site for cleavage by a proteolytic enzyme called sig-nal peptidase after translocation across the inner membrane. Chaperones such as SecB bind newly synthesized proteins to prevent folding and maintain a state that is competent for translocation (Fig. 18-9). Unlike most other chaperones (see Fig. 17-13), SecB does not require ATP hydrolysis for cycles of interac-tion with substrates. Hsp70 homologs (DnaK) have a secondary role in chaperoning precursors for translo-cation.
Translocation of many bacterial membrane and secreted proteins with cleavable signal sequences de-pend on the adenosine triphosphatase (ATPase) SecA. SecA binds proteins associated with SecB in the cytoplasm and targets the signal sequence to the Sec translocon. A system reconstituted from purified SecA, SecY, and SecE can translocate precursor proteins across lipid membranes in the presence of ATP. Remarkably, Archaea lack SecA, despite the fact that they depend on translocon components that are homologous to SecYE. Eukaryotes use SecA only for translocation into chloroplast thylakoids (Fig. 18-2).
Translocation Dependent on the Signal Recognition Particle
In eukaryotes, the signal recognition particle (SRP) is the adapter between signal sequences and the translocon of endoplasmic reticulum (see Fig. 20-3), but in bacteria, only a minority of integral membrane proteins and secreted proteins depend on SRP for targeting to the Sec translocon. Eukaryotic and archaeal SRPs consist of a 7S RNA and several proteins, whereas Escherichia coli SRP consists of a smaller 4.5S RNA and a single protein called Ffh (for “fifty-four homologue,” after its eukaryotic counterpart) (see Fig. 20-5). SRP binds Sec-signal sequences and signal-anchor sequences as they emerge from the ribosome. This interaction stops translation until SRP docks on the cytoplasmic surface of the inner membrane with its receptor FtsY and the Sec translocon. Resumption of translation drives the polypeptide through the translocon. See Chapter 20 for more details on SRP and eukaryotic cotranslational translocation.
Insertion of Proteins in the Outer Membrane of Gram-Negative Bacteria
Outer membrane proteins are synthesized in the cytoplasm and directed to the Sec translocon by signal sequences. The signal sequence is cleaved from the unfolded protein after crossing the inner membrane into the periplasm. No specific targeting signals are known for outer membrane proteins, so their localization likely depends on their tertiary structure. Individual protein subunits fold and then associate to form dimers and trimers (see Fig. 7-8C) before, or possibly after, insertion into the outer membrane. Several periplasmic assembly factors participate in protein folding, including enzymes that catalyze the isomerization of proline peptide bonds and oxidation/reduction of cysteine thiol groups.
Outer Membrane Autotransporter Pathway
Some proteins, including secreted proteolytic enzymes and toxins as well as membrane-anchored adhesins and invasins, hitch a ride to the cell surface on their own outer membrane transporters (Fig. 18-10A). These proteins are fused to a C-terminal β-domain that is thought to be similar to a porin (see Fig. 7-8). The protein uses the Sec pathway to cross the inner membrane and inserts into the outer membrane. The N-terminal functional domain then translocates across the outer membrane through its β-domain pore. An outer membrane protease releases toxins and proteases, whereas adhesins that follow this route remain on the surface attached to the β-domain.
Chaperone/Usher Pathway
Gram-negative bacteria use a novel mechanism, downstream of the Sec pathway, to transport and assemble pili on their outer surface. These appendages are involved with bacterial pathogenesis, including urinary tract infections. A periplasmic chaperone binds the pillus peptide and promotes folding. The pilus subunit is folded similar to an immunoglobulin (Ig) domain (see Fig. 3-13), but lacking the seventh β-strand. This exposes core hydrophobic residues. The chaperone consists of two immunoglobulin-like domains, one of which donates a strand to complete the immunoglobulin domain of the pilus subunit. The chaperone delivers a pilus subunit to an outer membrane translocon called usher (Fig. 18-10A). There, it transfers its bound subunit to the end of a growing chain of pilus subunits, all bound together, head to tail, by strands that complete the seven-strand β-sheet of the adjacent subunit. On the outer surface, the pilus subunits rearrange into a helical pilus. The assembly reaction is thought to provide the energy for translocation. The chaperone prevents premature assembly of the pilus.
Type II Secretion
Bacteria use an alternate route downstream of the Sec pathway to secrete other toxins and enzymes with cleaved signal sequences (Fig. 18-10A). At least a dozen protein subunits participate in this complicated pathway. The pore in the outer membrane is composed of a secretin, a protein with relatives that also participate in type III secretion, phage biogenesis, and formation of one type of pilus. The secretin pore is a ring of 12 to 14 subunits around a large but gated channel that is 5 to 10 nm in diameter.
Type IV Secretion
Bacteria secrete a few proteins using an apparatus similar to that used for DNA transfer between two bacteria during conjugation and for DNA injection into plant cells by Agrobacterium. DNA is transferred directly from the cytoplasm of one bacterium to the cytoplasm of another bacterium or plant cell. Proteins that are secreted by this pathway include pertussis toxin by Borditella pertussis and another toxin by Helicobacter pylori. This pathway starts with synthesis in the cytoplasm and translocation across the plasma membrane by the Sec translocon. If present, the signal sequence is cleaved before translocation across the outer membrane by the type IV secretion system (Fig. 18-10A).
Pathways Independent of the SEC Translocon
Type I ABC Transporters
Bacteria use ABC transporters (see Fig. 8-9) to secrete a small number of toxins (e.g., E. coli hemolysin), proteases, and lipases. C-terminal signal sequences of 30 to 60 residues target these proteins to the ABC transporter, the only component required for secretion by gram-positive bacteria. Gram-negative bacteria require not only a transporter in the inner membrane but also two proteins that form a continuous channel across the periplasm and outer membrane (Fig. 18-10B). ATP hydrolysis by the ABC transporter provides energy for translocation. Protein conduits across the periplasm and outer membrane engage ABC transporters presenting substrates for export and then disengage when translocation is complete. Genes for secreted proteins are generally in the same operon as the export machinery.
Flagellar and Type III Secretion Systems
The basal bodies of bacterial flagella transport flagellin subunits through a central pore that crosses both membranes (Fig. 18-10B) and extends the length of the flagellar shaft to the tip, where subunits add to the distal end (see Fig. 5-9). This flagellar pathway transports a few other proteins, including a phospholipase that contributes to the virulence of Yersinia, the cause of the black plague.
Pathogenic gram-negative bacteria, such as Yersinia, use the syringe-like type III apparatus, similar to a bacterial flagellum, to transport toxins from the cytoplasm into the medium or directly into target cells. In the target cell, these toxins disrupt cellular physiology, including formation of pores in target cell membranes. The type III secretion complex consists of about 20 different protein subunits. A complex base consisting of several protein rings spans the periplasm and both membranes. A polymer of a single protein forms a hollow needle up to 40 nm long for injection of toxins directly into target animal or plant cells.
Chen X, Schnell DJ. Protein import into chloroplasts. Trends Cell Biol. 1999;9:222-227.
Danese PN, Silhavy TJ. Targeting and assembly of periplasmic and outer-membrane proteins in E. coli. Annu Rev Genet. 1999;32:59-94.
Gutensohn M, Fan E, Frielingsdorf S, et al. Toc, Tic, Tat et al.: Structure and function of protein transport machines in chloroplasts. J Plant Physiol. 2006;163:333-347.
Keegstra K, Cline K. Protein import and routing systems of chloroplasts. Plant Cell. 1999;11:557-570.
Keegstra K, Froehlich JE. Protein import into chloroplasts. Curr Opin Cell Biol. 1999;2:471-476.
Koehler CM. New developments in mitochondrial assembly. Annu Rev Cell Devel Biol. 2004;20:309-335.
Koehler CM. The small TIM proteins and the twin Cx3C motif. Trends Biochem Sci. 2004;29:1-4.
Lazarow PB. Peroxisome biogenesis: Advances and conundrums. Curr Opin Cell Biol. 2003;15:489-497.
Nassoury N, Morse D. Protein targeting to the chloroplast of photosynthetic eukaryotes: Getting there is half the fun. Biochim Biophys Acta. 2005;1743:5-19.
Pfanner N. Protein sorting: Recognizing mitochondrial presequences. Curr Biol. 2000;10:R412-R415.
Pohlschroeder M, Dilks K, Hand NJ, Rose RW. Translocation of proteins across archaeal cytoplasmic membranes. FEMS Microbiol Rev. 2003;28:3-24.
Thanassi DG, Hultgren SJ. Multiple pathways allow protein secretion across the bacterial outer membrane. Curr Opin Cell Biol. 2000;12:420-430.
* This chapter was revised using material from the first edition written by William E. Balch, Ann L. Hubbard, J. David Castle, and Pat Shipman.
* PEX5 and PEX7 are import receptors for newly synthesized peroxisomal enzymes. Most other peroxins are also required for matrix enzyme import. Peroxins PEX3, PEX11, PEX16, and PEX19 are implicated in peroxisome membrane biogenesis rather than matrix protein import. AAA, AAA family of ATPases; CG, complementation group of patients with peroxisomal biogenesis disorders; PMP, peroxisomal membrane protein; SH3, Src-homology-3 domain; UBC, ubiquitin-conjugating enzyme. “Orphans” are novel proteins.