CHAPTER 44 Mitosis and Cytokinesi
Mitosis is the division of a somatic cell (a vegetative cell in yeast) into two daughter cells. The daughters are usually identical copies of the parent cell, but the process can be asymmetrical. For example, division of stem cells gives rise to one stem cell and another daughter cell that goes on to mature into a differentiated cell. See Box 41-1 for examples.
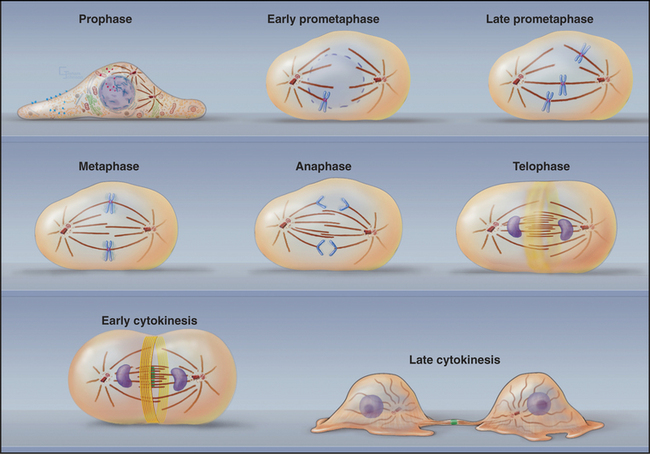
Traditionally, mitotic events are subdivided into six phases: prophase, prometaphase, metaphase, anaphase, telophase, and cytokinesis (Fig. 44-1). The dramatic reorganization of both the nucleus and cytoplasm during the mitotic phases is brought about by activation of a number of protein kinases, including Cdk1–cyclin B–p9 (abbreviated here as “Cdk1 kinase”; see Chapter 40). After activation by Cdc25 phosphatase, Cdk1 kinase accumulates in the nucleus, where it joins Cdk1–cyclin A, which was activated somewhat earlier (see Chapter 43). These two Cdk1 kinase complexes operate both as master controllers and as workhorses that directly phosphorylate many proteins whose functional and structural status is altered during mitosis.
Mitosis is an ancient eukaryotic process, and a number of variations emerged during evolution. Many single-celled eukaryotes, including yeast and slime molds, undergo a closed mitosis, in which spindle formation and chromosome segregation occur within an intact nuclear envelope to which the spindle poles are anchored. This chapter focuses on open mitosis, as used by most plants and animals, in which the nuclear envelope disassembles before the chromosomes segregate. Figure 44-2 summarizes some of the important events during the various mitotic phases.
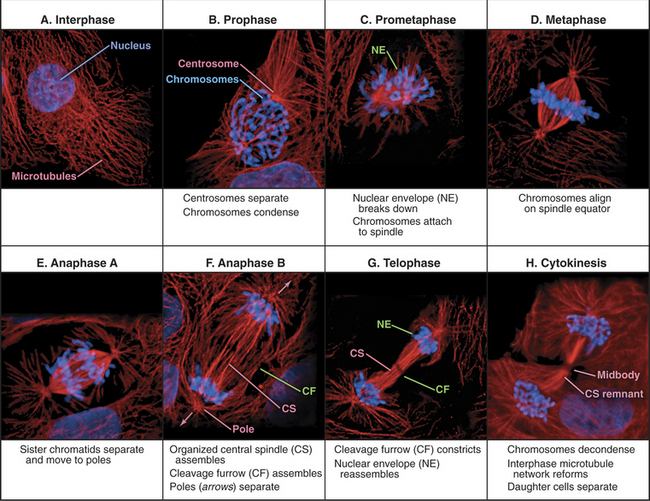
Figure 44-2 overview of the phases of mitosis and definition of the most important terms. a–c, Prophase–prometaphase: The Cdk1 kinases trigger condensation of replicated sister chromatids, disassembly of the nuclear envelope and Golgi, and a dramatic reorganization of the cytoskeleton. These changes abolish the barrier between the chromosomes and cytoplasm. As cytoplasmic microtubules contact the condensed chromosomes, they attach at the kinetochores (see Fig. 13-20). Interaction of motor proteins on the chromosomes with microtubules produces jostling movements that culminate with the chromosomes aligned at the midplane of a bipolar scaffolding of microtubules (the spindle). D–F, Metaphase–anaphase: Once all of the chromosomes achieve a bipolar attachment to the spindle, an inhibitory signal is switched off. This leads to activation of a proteolytic network that destroys proteins responsible for holding sister chromatids together and also inactivates Cdk1 by destroying its cyclin B cofactor (see Fig. 40-18). These changes trigger separation of the sister chromatids, which then move toward opposite spindle poles. G–H, Telophase–cytokinesis: Targeting of nuclear envelope components back to the surface of the chromatids subsequently leads to the re-formation of two daughter nuclei. In most cells, the two daughter nuclei and the surrounding cytoplasm are partitioned by cytokinesis following the contraction of an actin-myosin ring.
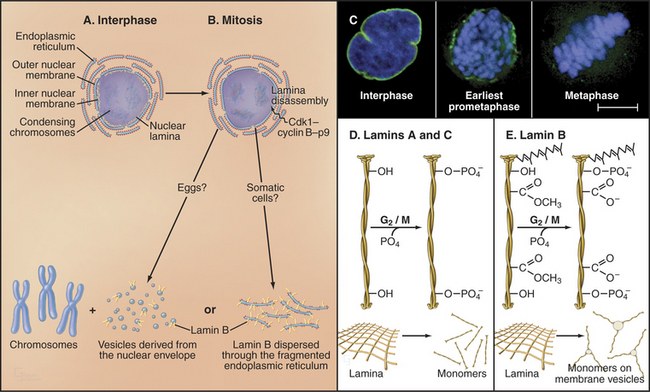
(Micrographs courtesy of William C. Earnshaw.)
Prophase
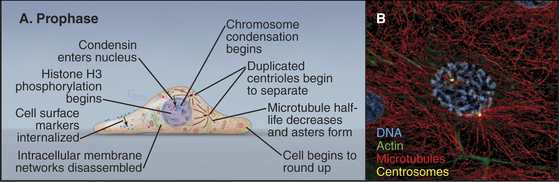
Prophase, the transition from G2 into mitosis, begins with the first visible condensation of the chromosomes and disassembly of the nucleolus (Fig. 44-3). In the cytoplasm, the interphase network of long microtubules centered on a single centrosome (see Fig. 34-17) is converted into two radial arrays of short microtubules called asters. Most types of intermediate filaments disassemble, the Golgi and endoplasmic reticulum fragment, and both endocytosis and exocytosis are curtailed.
Nuclear Changes in Prophase
Chromosome condensation, the landmark event at the onset of prophase, often begins in isolated patches of chromatin at the nuclear periphery. Later, chromosomes condense into two threads, termed sister chromatids, which are closely paired along their entire lengths. Although chromosome condensation was first observed more than a century ago, the biochemical mechanism remains a mystery. Protein kinases are thought to drive mitotic chromosome condensation by phosphorylating a number of the hundreds of proteins associated with mitotic chromosomes. The onset of condensation correlates with phosphorylation of histones H1 by Cdk1 kinase and H3 by Aurora-B protein kinase, and both are widely used as physiological markers for mitotic cells. However, chromosome condensation still occurs when both of these phosphorylation events are blocked. Possibly, some combination of histone modifications might provide a “code” that promotes chromatin condensation (see Fig. 13-3).
Two pentameric protein complexes, condensin I and II, are major constituents of mitotic chromosomes with an essential role in chromosome architecture (see Fig. 13-19). The two complexes share a pair of ABC ATPases, SMC2, and SMC4 (structural maintenance of chromosomes) but have two different sets of three auxiliary proteins. Condensin II enters the cell nucleus during prophase, where it is required for prophase chromosome condensation. However, chromosomes of vertebrate cells that lack condensin condense rapidly once the nuclear envelope breaks down at the onset of prometaphase, so condensin is not directly responsible for mitotic chromosome condensation. Chromosomes that lack condensin separate normally at the beginning of anaphase but appear to fall apart while moving toward the spindle poles. This appears to reflect defects in their underlying structure, and condensin is required for proteins of the chromosome scaffold to assemble properly (see Fig. 13-19). The molecular explanation for these effects is not known. In vitro, condensin complexes can promote coiling and compaction of DNA, but the significance of this is also unknown.
Cytoplasmic Changes in Prophase
Most of the cytoskeleton reorganizes during prophase. Most notably, the microtubule array changes from an extensive network permeating the cytoplasm into two dense, radial arrays of short, dynamic microtubules around the duplicated centrosomes (see Chapter 34). Each of these asters eventually becomes one pole of the mitotic spindle. During prophase, the two asters usually migrate apart across the surface of the nucle-ar envelope, signaling the start of spindle assembly (Fig. 44-3).
Mitotic microtubules behave like interphase microtubules in many ways (see Chapter 34). They are nucleated at their minus ends, they grow by addition of tubulin subunits at their free plus ends, and they undergo random catastrophes during which they rapidly shorten. To a large extent, the prophase changes in microtubule organization can be explained by two simple biochemical changes: (1) increased microtubule-nucleating activity of centrosomes and (2) altered dynamic instability properties of the microtubules (Table 44-1; also see Chapter 34). Interphase microtubules have a high probability of recovering from catastrophes, so they grow quite long. Mitotic microtubules grow more rapidly but exist only transiently. This is because when they undergo a catastrophe, they usually shorten all the way back to the centrosome, with little chance of rescue. These differences in dynamic instability can be reproduced in vitro in mitotic and interphase cellular extracts. They appear to arise, at least in part, from counterbalancing interactions between microtubule-associated proteins, which promote microtubule stability, and kinesin-13 (see Fig. 36-13), which promotes microtubule disas-sembly.
Table 44-1 COMPARISON OF MICROTUBULE DYNAMICS IN INTERPHASE AND MITOTIC NEWT LUNG CELLS
| Parameter | Interphase | Mitosis |
|---|---|---|
| Elongation rate | 7 μm/min | 14 μm/min |
| Elongation time before catastrophe | 71 s | 60 s |
| Shortening rate | 17 μm/min | 17 μm/min |
| Probability of rescue from catastrophe* | 0.046/s | 0 |
| Length | 100 μm | 14 μm |
* Most cellular microtubules grow constantly by addition of subunits to their free ends but occasionally stop growing and begin shrinking rapidly (a “catastrophe”). Unless shrinking is reversed (a “rescue”), the microtubule completely disappears.
(Data from Gliksman NR, Skib-bens RV, Salmon ED: How the transition frequencies of microtubule dynamic instability regulate microtubule dynamics in interphase and mitosis. Mol Biol Cell 4:1035–1050, 1993.)
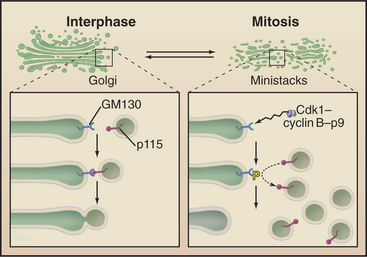
The Golgi apparatus and endoplasmic reticulum fragment or vesiculate during prophase (Fig. 44-4). In ad-dition, many membrane-mediated events, including fluid-phase pinocytosis, endocytosis, exocytosis, and intracellular sorting of membrane components (see Chapters 21 and 22), greatly decrease. Golgi disassembly is driven by several kinases, including Cdk1. The first step of the process is fragmentation of the Golgi into smaller mini-stacks. The second step is still being investigated. Some evidence argues that Cdk1 phosphorylation of key components prevents the fusion of transport vesicles back into Golgi stacks (see Chapter 21), the net result being that the Golgi buds apart into small vesicles. Other evidence suggests that an imbalance of vesicle flow between the Golgi and the endoplasmic reticulum results in the Golgi being absorbed into the ER during mitosis. Whatever the mechanism of its disassembly, Golgi reassembly begins again during anaphase, following inactivation of Cdk1 kinase. Some Golgi derived vesicles contribute to the plasma membrane during cleavage furrow ingression at the end of mitosis (see later).
Prometaphase
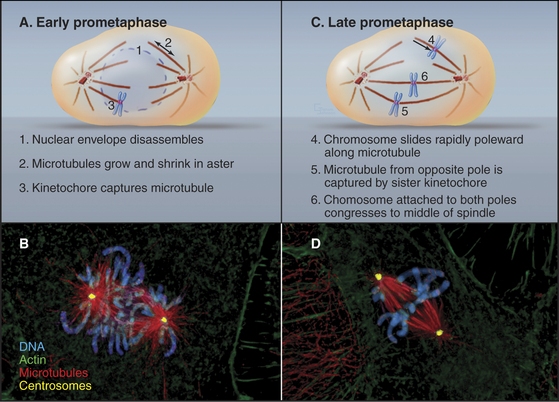
In cells that undergo an open mitosis, prometaphase begins abruptly with disassembly of the nuclear envelope (Fig. 44-5). Microtubules growing outward from the spindle poles penetrate holes in the nuclear envelope, make contact with the chromosomes, and attach to them at specialized structures called kinetochores (see Fig. 13-20). Interactions of the two opposing kinetochores of paired sister chromatids with microtubules from opposite poles of the spindle ultimately result in alignment of the chromosomes in a group midway between the poles. An important cell cycle checkpoint (see Chapter 40) known as the spindle checkpoint delays the onset of chromosome segregation until any attachment errors have been corrected and all chromosomes have achieved a bipolar attachment.
Nuclear Envelope Disassembly in Prometaphase
Nuclear envelope disassembly involves the removal of two membrane bilayers coupled with disassembly of the nuclear pores and the fibrous nuclear lamina meshwork that underlies the inner bilayer (Fig. 44-6). Protein phosphorylation triggers breakdown of the nuclear envelope but the critical targets and mechanisms are not entirely known. Phosphorylation of the nuclear lamins at two sites flanking the central coiled-coil causes the lamina network to disassemble into subunits and might contribute to disassembly of the envelope. Cdk1 kinase can phosphorylate these lamin residues in vitro, but other kinases might participate in vivo. Additionally, phosphorylation of nucleoporins leads to nuclear pore disassembly. This fenestrates the nuclear envelope, dissolving the barrier between nucleus and cytoplasm. Interaction between microtubules and dynein associated with the nuclear envelope may also rip holes in the envelope.
Nuclear envelope components are dispersed in the cytoplasm from prometaphase until telophase (Fig. 44-6), but the mechanism may differ in various cell types. In fertilized amphibian eggs, the nuclear membrane breaks up into small vesicles that disperse in the cytoplasm. In vertebrate somatic cells, the nuclear envelope may be absorbed into the endoplasmic reticulum, which remains as an extensive tubular network throughout mitosis. In both cases, lamin B remains associated with the dispersed nuclear envelope, whereas lamins A and C and many proteins of the nuclear pore complexes disperse as soluble subunits.
During prophase, kinetochores transform from nondescript balls of condensed chromatin into organized plaques on the surface of the chromosomes. By early prometaphase, the characteristic trilaminar disk structure (see Fig. 13-20) can be seen. Each sister chromatid has a kinetochore. Thus, sister kinetochores are located on opposite faces of the mitotic chromosome. Only one of the two sister kinetochores faces a given spindle pole at any one time.
Organization of the Mitotic Spindle
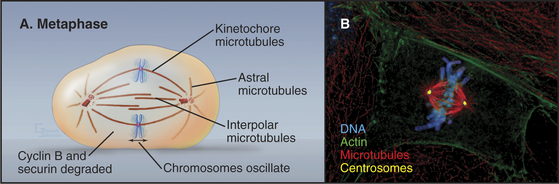
The mature metaphase spindle is a bilaterally symmetrical structure with centrally located chromosomes flanked by arrays of microtubules radiating from the poles (Fig. 44-7).
Three predominant classes of microtubules are present in the metaphase spindle (Fig. 44-12). Kinetochore microtubules have their plus ends embedded in the kinetochore and their minus ends at or near the spindle pole. They characteristically form bundles, called kinetochore fibers, which contain anywhere from 1 microtubule in the budding yeast to more than 200 microtubules in some higher plants. Each human kinetochore binds about 20 microtubules. Up to about 80% of the approximately 2200 spindle microtubules in humans may be present in kinetochore fibers. Interpolar microtubules are distributed throughout the body of the spindle and do not attach to kinetochores. Their minus ends may terminate near the pole but are not physically linked to it so that they appear to be free at both ends. Many interpolar microtubules penetrate between and through the chromosomes and extend for some distance beyond them. Thus, the central spindle contains a large number of interdigitated antiparallel microtubules. Tracking these spindle microtubules by electron microscopy has revealed a tendency for the interdigitated microtubules of opposite polarity to pack next to one another. During late anaphase, these antiparallel microtubules bundle to form a structure, called the central spindle, that appears to have important roles during cytokinesis. Astral microtubules project out from the poles and have a role in orienting the spindle in the cell through interactions with the cell cortex. All of the microtubules within each aster have the same polarity, with their minus ends proximal to the pole. Each unit of a spindle pole, with its associated kinetochore and interpolar and astral microtubules, is referred to as a half-spindle.
Spindle structure is largely determined by a combination of microtubule dynamics plus the action of at least seven different types of kinesins plus cytoplasmic dynein (see Chapter 36). These motors often work in opposition to one another. As a result, the spindle is a highly dynamic structure whose morphology changes as the balance of forces shifts between the various motors. For example, inactivating one or more kinesins with drugs or switching a temperature-sensitive mutant to the nonpermissive temperature can cause the spindle to collapse rapidly on itself. Consequently, chromosomal movements and changes in spindle morphology are complex processes that reflect both the dynamic growth and shrinkage of microtubules plus the net vectorial output of multiple antagonistic and synergistic motors. These various components interact; for example, force exerted by motors can influence the dynamic assembly/disassembly of microtubules.
Spindle Assembly
In metazoans, spindle assembly starts in prophase with the separation of the asters. In most cells, each aster is organized around a centrosome, consisting of a centriole pair and associated pericentriolar material. γ-Tubulin ring complexes in the pericentriolar material efficiently nucleate microtubules (see Fig. 34-16), so each aster acts as a microtubule organizing center. By the end of prophase, the spindle consists of two asters linked by a few interpolar microtubules. Cytoplasmic dynein at the cell cortex exerts an outward force separating the asters, whereas kinesin-14 motors (which move toward microtubule minus ends) on the interpolar microtubules exert a counterbalancing force holding the asters together.
This balance of forces changes when the nuclear envelope breaks down. Bipolar kinesin-5 motors are phosphorylated by Cdk1 kinase and concentrate in the central spindle, where they cross-link adjacent antiparallel interpolar microtubules. Kinesin-5 moves toward the plus ends of microtubules. If such a motor attaches to two adjacent antiparallel microtubules and begins to move, it will cause them to slide apart (Fig. 44-7). Thus, the action of kinesin-5 motors pushes the spindle poles apart. The two half-spindles do not separate because they are physically linked via the chromosomes, with sister kinetochores attached to opposite spindle poles.
Also at this time, the asters mature into focused spindle poles. The pericentriolar material efficiently nucleates the assembly of new microtubules with their minus ends at the pole. In addition, cytoplasmic dynein transports free microtubules that are nucleated throughout the cytoplasm to the centrosome along astral microtubules for incorporation into the spindle. The focused microtubule array at the pole forms partly owing to the tethering of microtubules by centrosomes, and partly due to the concerted action of various motors and microtubule cross-linking proteins such as nuclear mitotic apparatus protein (NuMA). NuMA is released from the nucleus on nuclear envelope breakdown, and it accumulates near the poles at the minus ends of microtubules.
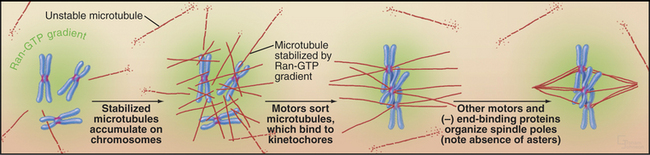
In large cells that lack centrosomes, such as eggs, spindle formation depends on an alternative pathway that is also active in cells with centrosomes (Fig. 44-8). Chromosomes stabilize nearby microtubules, which are then organized into a bipolar spindle by motor proteins and NuMA. This spindle assembly pathway involves importin a and b, two proteins that direct traffic through nuclear pores during interphase (see Fig. 16-14). Importin a and b inhibit mitotic spindle formation by sequestering several essential proteins, including NuMA. Chromosomes counteract this by releasing spindle assembly factors such as NuMA from importin a and b. The mechanism depends on the association of the GTP exchange factor RCC1 (Ran-GEF in Fig. 44-17) with chromosomes. RCC1 produces a local gradient of the active GTPase Ran-GTP, which dissociates NuMA from importin a and b just as it does during protein import into the nucleus. In this case, however, the net result is microtubule stabilization and assembly of the mitotic spindle.
Chromosome Attachment to the Spindle
Dynamic microtubules of prometaphase asters scan the cytoplasm searching for both binding sites that will capture and stabilize their distal plus ends and for other components, including free microtubules. Breakdown of the nuclear envelope makes the condensed chromosomes accessible to the microtubules. Chance encounters with kinetochores during cycles of growth result in the plus ends of microtubules being captured by the kinetochore. Capture probably involves the nine-component KMN complex, which contains two components capable of binding weakly to microtubules. One of these, the helical Ndc80 complex (see Fig. 13-21) binds along the sides of microtubules forming fine hairs visible in the electron microscope. Other members of the complex provide an anchoring site for Ncd80 in the kinetochore. Captured microtubules are about five-fold less likely to depolymerize catastrophically than free microtubules. When catastrophes do occur, the microtubules depolymerize back to the pole, recycling tubu-lin subunits for incorporation into other, growing microtubules.
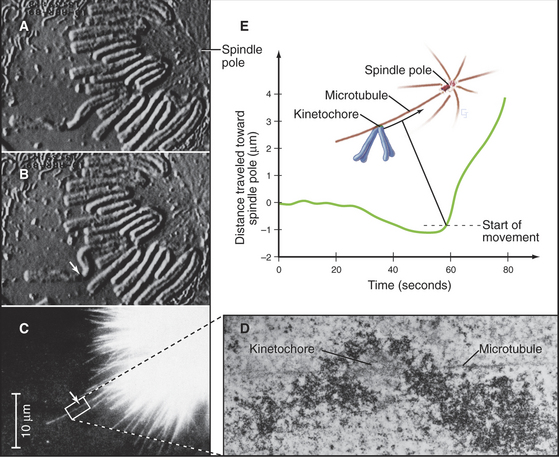
Initial attachment of a chromosome to a microtubule often involves a lateral interaction between the corona region of the kinetochore (see Fig. 13-20) and the side of a microtubule (Fig. 44-9). Cytoplasmic dynein then slides the chromosome rapidly along the microtubule toward the pole. These steps were first seen in animal cells, and a similar pattern of chromosome attachment and movement also occurs in budding yeast cells.
Capture of the first microtubule by a kinetochore causes the chromosome to move initially toward the spindle pole from which that microtubule originated. Historically, it has been thought that subsequent cap-ture of a microtubule emanating from the opposite spindle pole by the sister kinetochore provides a counterforce that tugs the chromosome in the opposite direction. This bipolar attachment produces a balance of opposing forces that, together with the action of kinesin family motor proteins distributed along the chromosome arms, results in the gradual movement of the chromosome toward a point midway between the spindle poles. These movements are accompanied by coordinated shrinkage of the microtubules at the leading kinetochore and growth of microtubules at the trailing kinetochore. More recently, it has been shown that chromosomes attached to only one spindle pole (mono-oriented) can move toward the spindle equator if the unattached kinetochore associates with the kinetochore fiber of a chromosome that has already become aligned at the spindle equator. In this case, the mono-oriented chromosome glides away from the pole to which it is attached toward the spindle midzone, where it is more likely to capture microtubules emanating from the opposite pole. This motion of one chromosome along the kinetochore fiber of another chromosome requires the kinesin-7 motor CENP-E, which is associated with the kinetochore of the moving chromosome.
Correcting Errors in Chromosome Attachment to the Spindle
Attachment of both sister kinetochores to a single spindle pole is rare, since sister kinetochores are positioned on opposite faces of the chromosome (see Fig. 13-20). When it occurs, one or both kinetochores must detach for the chromosome to achieve a bipolar orientation. Chromosome attachment to opposite spindle poles is more stable than attachment to a single pole, because the tension generated by bipolar attachment (where forces pull a chromosome simultaneously toward opposite spindle poles) preferentially stabilizes microtubule connections to both kinetochores. It follows that the attachment of a single kinetochore to both spindle poles is more insidious, as that kinetochore is under tension, and the attachments are therefore stable. In fact, attachment of a single kinetochore to both poles seems to be the most common cause of chromosome segregation errors in cultured mammalian cells.
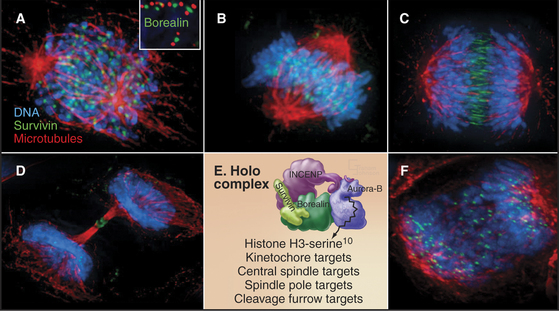
Both of these chromosome attachment errors are corrected through the action of Aurora B protein kinase, a member of the chromosomal passenger complex, along with inner centromere protein (INCENP), surviving, and borealin (Fig. 44-10). The other subunits target Aurora B to its various points of action during mitosis and regulate the kinase activity. The complex concentrates in the inner centromeres (the heterochromatin beneath and between the two sister kinetochores) during prometaphase and metaphase. As sister chromatids separate at anaphase, the complex moves to the overlapping interpolar microtubules of the central spindle and to the cell cortex, where the cleavage furrow will form, ultimately winding up in the intercellular bridge during cytokinesis. The chromosomal passenger complex is required to complete cytokinesis but also contributes to the correction of chromosome attachment errors and to the operation of the checkpoint that delays the cell cycle in response to those errors (Fig. 44-10).
Aurora B responds to tension on kinetochores to correct chromosome attachment errors. It phosphorylates both Ndc80 and the Dam1 complex, which are involved in microtubule binding to the kinetochore (see Fig. 13-21). Aurora B phosphorylation strongly inhibits Ndc80 binding to microtubules, and this may signal the kinetochore to let go of the attached microtubule. When a chromosome is correctly attached to both spindle poles, tension may stretch the kinetochore away from the chromosomal passenger complex buried in the chromatin beneath. This might stabilize the chromosome-microtubule interaction by preventing the kinase from phosphorylating the kinetochore.
Finding Time to Fix Chromosome Attachment Errors: The Spindle Checkpoint
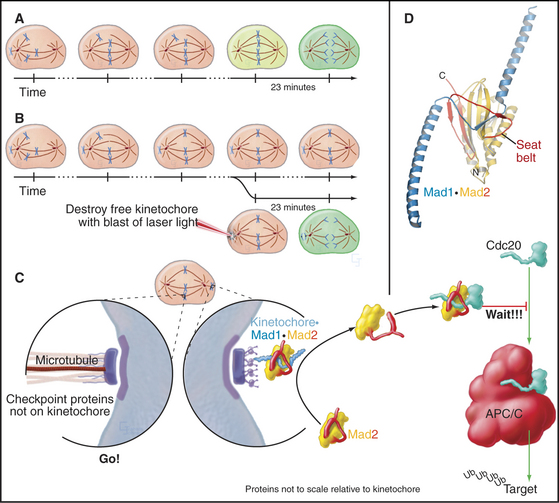
Segregation of replicated chromosomes into daughter cells is extremely accurate. For example, budding yeasts lose a chromosome only once in 100,000 cell divisions. Perhaps surprisingly, the frequency of chromosome loss may be 20-fold to 400-fold higher for human cells grown in culture. To achieve even this level of accuracy, most cells must delay entry into anaphase until all chromosomes are attached to both poles of the mitotic spindle. This delay is caused by a cellular quality control pathway or checkpoint—the spindle checkpoint—that senses the completion of chromosome alignment at metaphase (Fig. 44-11). This nomenclature is nearly universal, but it is worth noting that this checkpoint actually monitors kinetochore activity rather than spindle structure. The spindle checkpoint differs from the DNA damage checkpoints in that its default setting is “on” as cells enter mitosis. It shuts off only when every chromosome is properly attached to the spindle.
The target of the spindle checkpoint is Cdc20. This protein is thought to be a substrate recognition factor for the APC/C, a ubiquitin-protein ligase (E3 enzyme; see Chapter 23 and Fig. 44-16) that marks target proteins for destruction by proteasomes by decorating them with ubiquitin. Key APC/C substrates include cyclin B and a protein called securin, an inhibitor of the enzyme that triggers separation of sister chromatids at anaphase (see Fig. 44-16). Despite intense study, the mechanistic details are still debated.
Mad1 protein binds to kinetochores that are not properly attached to the spindle (Fig. 44-11). The best guess is that Mad1 then binds Mad2. A loop on Mad2 wraps around Mad1 like a safety belt to make a complex that resides stably at kinetochores. This complex can bind additional Mad2 molecules and convert them to a conformation activated for Cdc20 binding. The “primed” Mad2 molecules are then released either on their own or as part of a “mitotic checkpoint complex”; they bind Cdc20, and the APC/C is inhibited. As each chromosome becomes attached to both poles of the spindle, its inhibitory signals are removed. When the last chromosome has achieved a proper attachment, the last source of inhibitory Mad2 complexes is extinguished, and mitosis can proceed.
In metazoans, two of the checkpoint components are protein kinases, and one of these, BubR1, may be involved in sensing the quality of kinetochore attachments to the spindle. When the nuclear envelope breaks down at prometaphase, the COOH-terminus of the CENP-E kinesin binds to BubR1 located in the outer plate of the kinetochore. This interaction stimulates the BubR1 kinase activity, a possible early step in activating the checkpoint. Subsequent microtubule binding by CENP-E then shuts the kinase off again. Thus, CENP-E may function in recognition of microtubule binding by the kinetochore. The yeast homolog of BubR1 is not a kinase; furthermore, yeasts lack CENP-E. Therefore, how they sense microtubule attachment to kinetochores remains mysterious.
Metaphase
When all of the chromosomes have attained bipolar orientations (attached to both spindle poles) and moved to positions roughly midway between the two spindle poles, the cell is said to be in metaphase (Fig. 44-12). The compact grouping of chromosomes at the middle of the spindle is referred to as the metaphase plate. Destruction of cyclin B and securin (see Fig. 44-16) triggered by the APC/C begins as soon as the last chromosome achieves a bipolar orientation and continues throughout metaphase. Loss of securin is the signal for the separation of sister chromatids, the first sign of anaphase onset. Degradation of cyclin A, which with Cdk1 had an important role in triggering the entry into mitosis, begins earlier, at the entry into prometaphase, and is completed by mid-metaphase.
Microtubule Flux within the Metaphase Spindle
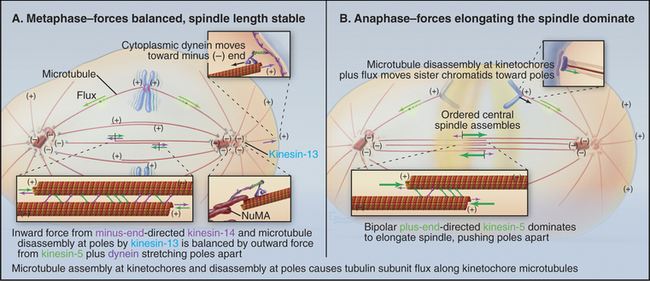
Although the average length of the kinetochore microtubules is roughly constant during metaphase, the microtubules change continuously in three ways. First, there is constant net addition of new tubulin subunits (about 10 subunits per second) to the plus end of the microtubules, where they are attached to the kinetochore. Second, a comparable number of tubulin subunits is continuously lost from the minus end of the kinetochore tubules at the spindle poles. Therefore, tubulin subunits slowly migrate through kinetochore microtubules from the kinetochore to the pole (Fig. 44-13). This subunit flux or treadmilling is caused by microtubule depolymerization at the poles driven by kinesin-13 family members. Third, all microtubules attached to each kinetochore change coordinately in length during chromosomal oscillations (see next section).
Chromosome Oscillations during Metaphase
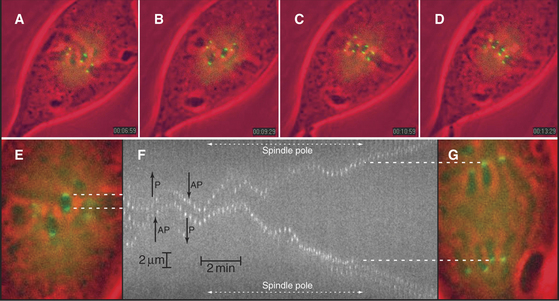
In many cells, even though chromosomes remain, on average, balanced at the middle of the spindle, they jostle one another and undergo numerous small excursions toward one pole or the other throughout metaphase (Fig. 44-14). These oscillations are slow, about 1.5μm/minute (gain or loss of about 40 tubulin subunits per microtubule per second), perhaps because they require the simultaneous shortening or lengthening of the approximately 20 microtubules attached to each kinetochore in vertebrate cells. The oscillatory movements reverse every few minutes. Both active movements by motor proteins and fluctuations in the length of kinetochore microtubules contribute to chromosome oscillations during metaphase, but their relative contributions may vary in different cell types. Although the movements of paired sister chromatids are usually coordinated, uncoordinated movements can stretch or compress centromeric chromatin. Thus, although each kinetochore can act independently, some mechanism (perhaps tension sensors in the kinetochore) usually coordinates their actions.
Anaphase
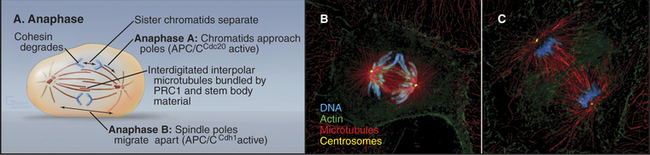
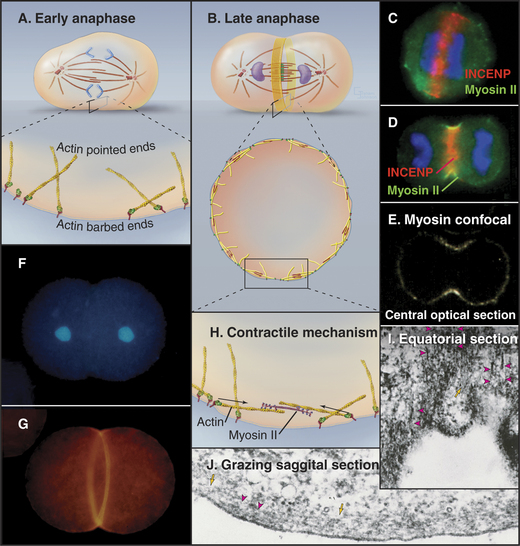
The separation of sister chromatids at the onset of anaphase is one of the most dramatic events of the entire cell cycle (Fig. 44-15). Sister chromatids move to opposite spindle poles (anaphase A), and the poles move apart (anaphase B). Anaphase is also the time when the mitotic spindle activates the cell cortex in preparation for cytokinesis.
The dramatic physiological transition of the cytoplasm at anaphase is triggered by the action of two forms of the APC/C and degradation of key proteins. Anaphase A follows activation of APC/CCdc20 (i.e., the APC/C with substrate specificity determined by Cdc20) during metaphase, which targets cyclin B for degradation and causes Cdk activity to fall (see Fig. 40-18). Anaphase B depends on activation of APC/CCdh1, which forms only when Cdk kinase activity has dropped. This is because Cdh1 phosphorylated by Cdk1 kinase cannot bind to the APC/C. APC/CCdh1 targets polypeptides whose destruction by the proteasome is required for the cell to exit from mitosis and return to interphase.
Biochemical Mechanism of Sister Chromatid Separation
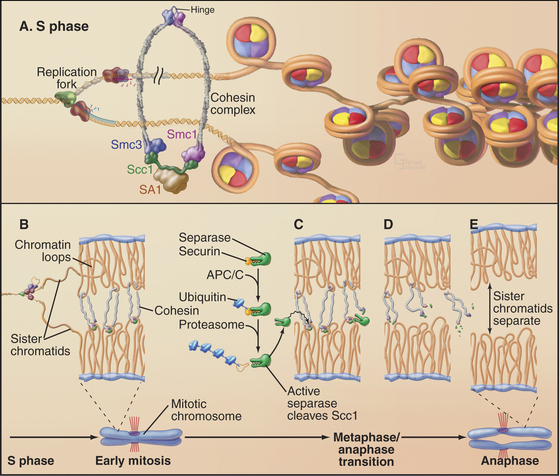
Studies of the budding yeast led to a major breakthrough by revealing three factors that regulate sister chromatid separation: a protein complex known as cohesin, a protease known as separase, and an inhibitor of the protease known as securin (Fig. 44-16). This system is conserved from yeast to human.
Cohesin is a complex of four proteins that resembles the condensin complex (see Fig. 13-19). Like condensin, cohesin has two large subunits from the SMC (structural maintenance of chromosomes) family. These proteins, SMC1 and SMC3, are complexed with proteins called Scc1 (which has other names omitted here for simplicity) and Scc3. Additional proteins are required for the stable loading of this complex onto DNA. Cells with mutations in cohesin components separate sister chromatids prematurely in mitosis, resulting in chaotic chromosome missegregation. This system is very ancient; an SMC-related protein is required for orderly chromosome segregation in bacteria.
Exactly how cohesin holds sister chromatids together is unknown, but a variety of evidence suggests that it could form a ring with a diameter of 40 nm, large enough to encircle two sister chromatids like a lasso. In yeast, the complex functions only if it binds chromosomes during DNA replication, and one factor involved is a specialized form of the RFC complex that loads PCNA rings onto DNA (see Fig. 42-11). Cohesin accumulates at preferred sites on the chromosomes, often near centromeres in budding yeast or in regions of heterochromatin in fission yeast. In vertebrates, most cohesin dissociates from the chromosome arms by late metaphase, owing to the action of protein kinases such as Plk1 and Aurora B, but some remains associated with heterochromatin flanking centromeres until the onset of anaphase.
Cleavage of two key proteins triggers sister chromatid separation at anaphase. The first of these, securin, is an inhibitor of the separase protease. After the last chromosome forms a bipolar attachment to the spindle, the spindle checkpoint is switched off. This allows APC/CCdc20 to tag securin with ubiquitin, leading to its destruction by proteasomes throughout metaphase. When securin levels fall below a critical threshold, separase is unleashed to cleave the Scc1 subunit of cohesin. Cleavage of Scc1 either breaks or disassembles the cohesin ring, allowing the sister chromatids to separate, thereby triggering the onset of anaphase. Phosphorylation of the separase cleavage site on Scc1 by a protein kinase can increase the cleavage of Scc1 by separase. Thus, the proteolysis of Scc1 is integrated and coordinated with the activities of the various mitotic kinases (see Chapter 40).
Human securin is overexpressed in some pituitary tumors, and the protein can act as an oncogene in cultured cells (see Fig. 41-10). Overexpression of securin may disrupt the timing of chromosome segregation, leading to chromosome loss and ultimately contributing to cancer progression.
Mitotic Spindle Dynamics and Chromosome Movement during Anaphase
Anaphase is dominated by the orderly movement of sister chromatids to opposite spindle poles brought about by the combined action of motor proteins and changes in the length of microtubules. Anaphase chromosome movements occur in two phases (Fig. 44-15). Anaphase A, the movement of the sister chromatids to the spindle poles, requires a shortening of the kinetochore fibers. During anaphase B, the spindle elongates, pushing the spindle poles apart. The poles separate partially because of interactions between the antiparallel interpolar microtubules of the central spindle and partially because of intrinsic motility of the asters. Most cells use both components of anaphase, but one component may be strongly exaggerated in relation to the other.
Microtubule disassembly on its own can move chromosomes (see Fig. 37-8). Energy for this movement comes from hydrolysis of GTP bound to assembled tubulin, which is stored in the conformation of the tubulin subunits. Chromosomes appear to use motor molecules such as cytoplasmic dynein in the kinetochore corona to hold onto disassembling microtubules. These motors must “run” toward the poles without losing their grip as the microtubules disassemble behind them. In addition, at yeast kinetochores the Dam1 ring (green in Fig. 44-21) can remain associated with disassembling microtubules. Other kinesin “motors” influence the dynamic instability of the spindle microtubules. Members of the kinesin-13 class, which encircle microtubules near kinetochores and at spindle poles, use ATP hydrolysis to promote microtubule disassembly rather than movement.
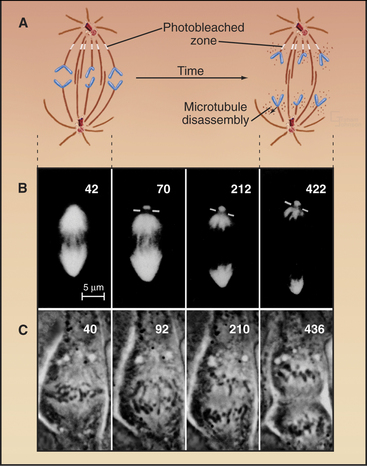
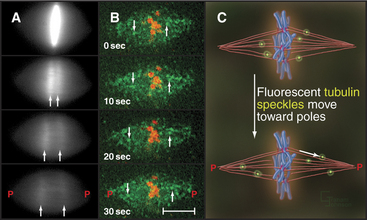
Anaphase A chromosome movement involves a combination of microtubule shortening and translocation of the microtubule lattice due to flux of tubulin subunits (Fig. 44-13). The contributions of the two mechanisms vary among different cell types. When living vertebrate cells are injected with fluorescently labeled tubulin subunits, the spindle becomes fluorescent (Fig. 44-17). If a laser is used to bleach a narrow zone in the fluorescent tubulin across the spindle between the chromosomes and the pole early in anaphase, the chromosomes ap-proach the bleached zone much faster than the bleached zone approaches the spindle pole. This shows that the chromosomes “eat” their way along the kinetochore microtubules toward the pole. In these cells, subunit flux accounts for only 20% to 30% of chromosome movement during anaphase A, and this flux is dispensable for chromosome movement. In Drosophila embryos, in which subunit flux accounts for about 90% of anaphase A chromosome movement, the chromosomes catch up with a marked region of the kinetochore fiber slowly, if at all.
Anaphase B appears to be triggered by the inactivation of the minus end-directed kinesin-14 motors, so that all of the net motor force favors spindle elongation. Three factors contribute to overall lengthening of the spindle: sliding apart of the interdigitated half-spindles, microtubule growth, and intrinsic motility of the poles themselves (Fig. 44-7). During the latter stages of anaphase B, the spindle poles, with their attached kinetochore microtubules, appear to move away from the interpolar microtubules as the spindle lengthens. This astral movement involves interaction of the astral microtubules with cytoplasmic dynein molecules anchored in the cortical cytoplasm.
Anaphase B spindle elongation is accompanied by reorganization of the interpolar microtubules into a highly organized central spindle between the separating chromatids (Fig. 44-15). Within the central spindle, an amorphous dense material called stem body matrix stabilizes bundles of antiparallel microtubules and holds together the two interdigitated half-spindles. Proteins concentrated in the central spindle help to regulate cytokinesis. One key factor, PRC1 (protein regulated in cytokinesis), is inactive when phosphorylated by Cdk kinase and functions only during anaphase when Cdk activity declines. PRC1 directs the binding of several kinesins to the central spindle. Each kinesin appears to target a specific protein kinase, such as Aurora B, to a particular domain of the central spindle, where phosphorylation of key substrates then regulates spindle elongation and cytokinesis.
Telophase
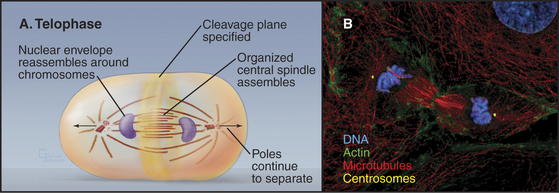
During telophase, the nuclear envelope re-forms on the surface of the separated sister chromatids, which typically cluster in a dense mass near the spindle poles (Fig. 44-18). Some further anaphase B movement may still occur, but the most dramatic change in cellular structure at this time is the constriction of the cleavage furrow and subsequent cytokinesis.
Reassembly of the Nuclear Envelope
Nuclear envelope reassembly begins during anaphase and is completed during telophase (Fig. 44-19). As in spindle assembly, Ran-GTP promotes early steps of nuclear envelope assembly by releasing near the surface of the chromosomes key components that were sequestered by importin b. Most of these factors are not yet identified, but they include several nuclear pore components.
The mechanism of nuclear envelope reassembly is debated, in part because the fate of the nuclear membrane during mitosis is unclear. If the nuclear envelope disassembles to discrete vesicles as it does in eggs, then envelope reassembly is a classic membrane-sorting problem (see Fig. 21-12) that requires the fusion of membrane vesicles. In eggs, Ran-GTP and unknown factors direct the fusion of at least two or three discrete populations of membrane vesicles to re-form the nuclear envelope. On the other hand, if the nuclear membrane is absorbed into the endoplasmic reticulum during mitosis, as in vertebrate somatic cells, then reassembly involves lateral movements of membrane components within the membrane network and their stabilization at preferred binding sites at the periphery of the chromosomes.
Lamin subunits disassembled in prophase are recycled to re-form the nuclear envelope at the end of mitosis. Reassembly of the nuclear lamina is triggered by removal of mitosis-specific phosphate groups and methyl-esterification of several COOH side chains on lamin B (Fig. 44-6). B-type lamins are among the earliest components of the nuclear envelope to target to the surface of the chromosomes during mid-anaphase. Either at this time or shortly thereafter, other proteins associated with the inner nuclear membrane, including BAF, LAP2, and lamin B receptor (see Fig. 14-8), join the forming envelope. Lamin A enters the re-forming nucleus later during telophase, after the reassembly of nuclear pore complexes and reestablishment of nuclear import pathways. Its assembly into the peripheral lamina occurs slowly over a period of several hours in the G1 phase. Transport of lamins through nuclear pores appears to be essential for nuclear reassembly. If lamin transport is prevented, chromosomes remain highly condensed following cytokinesis, and the cells fail to reenter the next S phase.
Cytokinesis
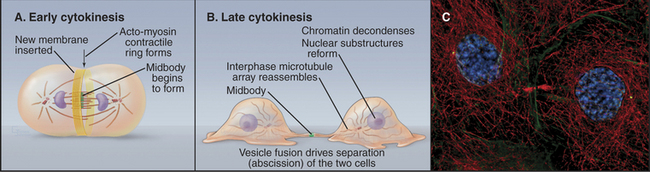
Cytokinesis is the process that divides a mitotic cell into two daughter cells (Fig. 44-20). Cytokinesis involves a number of mechanistically distinct events. These include signaling to specify the cleavage plane (Fig. 44-21), assembly and regulation of the contractile apparatus, specific alterations (including targeted growth) of the cell membrane, and the final separation (abscission) of the two daughter cells.
In animals, protozoa, and most fungi a contractile ring of actin filaments and myosin-II separates daughter cells at the end of mitosis. Myosin-II pulls on the ring of actin filaments, applying tension to the plasma membrane, much like contraction of smooth muscle (see Figs. 39-20 and 39-21). Because the contractile ring is confined to a narrow band of cortex around the equator, it forms a cleavage furrow, constricting the plasma membrane locally and pinching the cell in two like a purse string (Fig. 44-20). Signals from the mitotic spindle and cell cycle machinery control the position of this ring and the timing of its constriction.
Protozoa, animals, fungi, and plants use an evolutionarily conserved set of components to implement different strategies to separate daughter cells. For example, both fission yeast and fruit fly cells use signals from polo kinase, a Rho GTPase, and a GTPase-activating protein (GAP) to direct the assembly of a contractile ring of actin, myosin-II, and other conserved components, in spite of the fact that the yeast has a closed mitosis and the flies have an open mitosis. Plants divide by targeted fusion of membrane vesicles to build a new cell wall rather than constricting a cleavage furrow as animals do (Box 44-1 and Fig. 44-22A). However, the final abscission of animal cells also involves targeted fusion of vesicles, and the process is controlled by syntaxins in plants, animal cells, and fungi. Differences between various model organisms may be apparent or real, but they have certainly complicated the quest for a unified model for cytokinesis. Cytokinesis in prokaryotes is genuinely different, since completely different proteins are involved (Fig. 44-22B).
BOX 44-1 Variations on a Theme: Cytokinesis in Plants and Bacteria
Plants
Chromosome segregation is similar in plants and animals, but cytokinesis is very different (Fig. 44-2A). Plants lack centrosomes, and during interphase, microtubules radiate out from the surface of the cell nucleus in all directions. In mitosis, the spindle does not focus to sharp poles at metaphase; instead, it assumes a barrel shape with flat poles. Early in mitosis, a band of microtubules and actin filaments forms around the equator of the cell adjacent to the nucleus. This so-called preprophase band disassembles as cells enter prometaphase. Because the entire cell cortex is covered by a meshwork of actin filaments, disassembly of the preprophase band actually leaves an actin-poor zone in a ring where cytokinesis will ultimately occur. This is called the cortical division site. In late anaphase, two nonoverlapping, antiparallel arrays of microtubules form over the central spindle. This structure, the phragmoplast, gradually ex-pands laterally until it makes a mirror-symmetric double disk of short microtubules with their plus ends abutting the plane of cell cleavage. In addition to microtubules, the phragmoplast contains actin filaments and vesicles derived from the Golgi apparatus and endoplasmic reticulum. The Golgi vesicles, containing cell wall materials (see Fig. 32-12), move along phragmoplast microtubules to the equator, where they fuse, forming a membrane network that will become the new plasma membrane and laying down the material that will become the new cell wall. As the zone of newly deposited membrane expands radially, the ring of microtubules surrounding it similarly expands. Eventually, the new membrane reaches the lateral cell periphery, and fusion with the plasma membrane separates the two daughter cells. The cortical division site, not the spindle, determines the site of cleavage. This was shown by centrifuging mitotic cells to displace the spindle from the central location where it initially formed. Late in mitosis, the phragmoplast formed at the midzone of the displaced spindle, but this phragmoplast then migrated to the plane of the preprophase band, where cytokinesis occurred.
Bacteria
The strategy for cytokinesis in bacteria is similar to that in animal cells (Fig. 44-22B), but the molecules are completely different. Most bacterial cells cleave as a result of constriction of a ring of the FtsZ protein (filamentous temperature-sensitive; mutants in fts genes cannot divide and make long filaments on cells). This is called the Z ring. FtsZ is the prokaryotic homolog of eukaryotic tubulins, but it assembles into filaments rather than tubules. As for tubulins (see Fig. 34-4), FtsZ polymerization requires bound GTP and hydrolysis of this GTP destabilizes the polymers.
The Z ring is positioned at the cell equator of Escherichia coli by the action of three gene products: MinC, MinD, and MinE (minicell mutants divide at inappropriate locations and give birth to tiny cells). MinD is an enzyme that recruits MinC to the cell cortex, where it inhibits Z-ring formation. MinE is an antagonist of MinC/MinD action. This system works in a truly remarkable way. MinE forms a ring at the cell equator that migrates along the inner surface of the cell membrane until it reaches the end of the cell, at which point it disassembles. The ring then re-forms in the center of the cell and sweeps toward the other end of the cell. As it moves, MinE inactivates the MinC/MinD inhibitory complex on the cell cortex. The inhibitory complex rapidly reestablishes itself on the cell cortex behind the moving MinE ring. It takes about two minutes for each sweep of the MinE ring along half of the cell, and this cycle is repeated continuously until the FtsZ ring assembles at the cell center. No one knows how MinE and FtsZ locate the center of the cell. Bacillus subtilis uses an alternative mechanism to position the Z ring for cytokinesis. Interestingly, chloroplasts use a similar system for their division, and FtsZ has been detected in mitochondria of certain primitive eukaryotes. Mitochondria of higher eukaryotes appear to use another GTPase, dynamin, for a similar cleavage mechanism (see Chapter 19, under the section titled “Biogenesis of Mitochondria”).
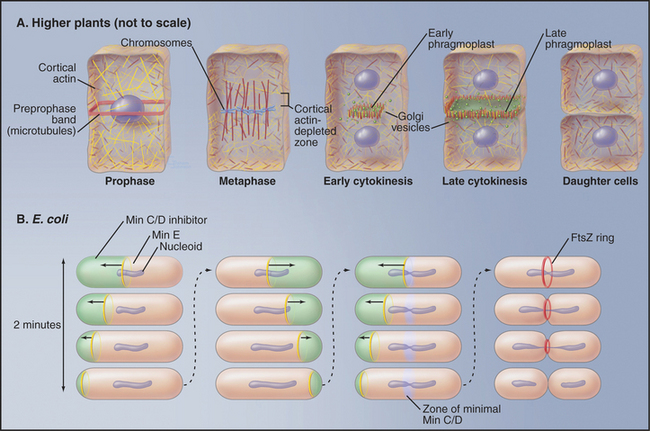
Figure 44-22 regulation of cytokinesis. A, Higher plants. B, Escherichia coli. See the text for details.
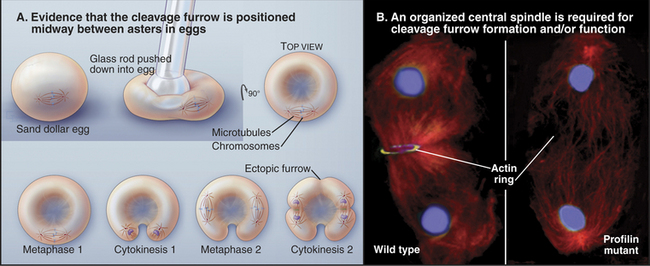
Signals Regulating the Position of the Cleavage Furrow
Elegant experimental data from classic studies on fertilized echinoderm eggs suggest that a cleavage stimulus, emitted by the mitotic spindle, specifies the position of the cleavage furrow midway between the poles and perpendicular to the long axis of the spindle, thereby ensuring that the cleavage process separates the daughter nuclei (Fig. 44-21). In fertilized eggs, the poles, with their large astral arrays of microtubules, are regarded as the source of the cleavage stimulus, as furrows can be induced to form midway between two poles, even when no chromosomes are present. In addition, a signal emitted by the bundled microtubules of the central spindle appears to modulate the behavior of the furrow signaled by the poles.
Assembly and Regulation of the Contractile Ring
Exposure of the cell cortex to the cleavage stimulus culminates in the assembly of a contractile ring consisting of a very thin (0.1 to 0.2μm) array of actin filaments attached to the plasma membrane at many sites around the equator (Fig. 44-23). Polymerization of the actin filaments depends on formins (see Fig. 33-12). Small, bipolar filaments of myosin-II are interdigitated with actin filaments. The plasma membrane adjacent to this actin-myosin ring undergoes alterations in its lipid composition that are important for the function of the contractile ring.
Membrane furrowing requires actin and the motor activity of myosin-II (see Fig. 36-5). In animals, the small GTPase RhoA regulates actin polymerization by formins as well as constriction of the ring. Many other proteins are required for cytokinesis to go to completion. In their absence, furrowing begins, but the cleavage furrows ultimately regress, producing binucleated cells. This large class of proteins includes anillin, the chromosomal passenger proteins (Aurora-B kinase and its associated subunits [Fig. 44-10]) and a complex of Rho-GAP with a kinesin-6, among many others. Anillin helps to keep active myosin-II focused into an organized contractile ring throughout cytokinesis.
Fission yeast assemble a contractile ring along a well-defined pathway by recruiting proteins from cytoplasmic pools (Fig. 44-24). Polo kinase releases an anillin-like protein from the nucleus to mark the cortex in the middle of the cell and recruit myosin-II and a formin. Profilin activates the formin to polymerize actin filaments. Myosin-II pulls the actin filaments together into a ring around the equator of the cell (Fig. 44-23).
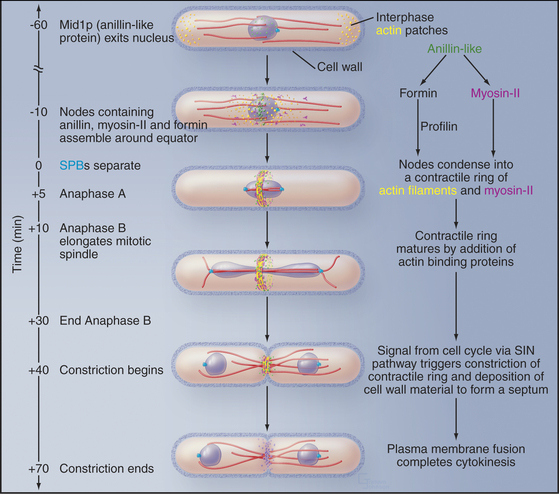
Figure 44-24 Cytokinesis in fission yeast Schizosaccharomyces pombe. During interphase, microtubules (red) position the nucleus in the middle of the cell. Actin filaments concentrate in small patches (yellow) in the cortex at the two growing ends of the cell (see Fig. 33-1). The mitotic spindle is inside the nucleus, as the nuclear membrane does not break down during mitosis. As the cell enters mitosis, an anillin-like protein moves from the nucleus to the equatorial cortex, where it sets up nodes of proteins, including myosin-II and a formin. The formin grows actin filaments (yellow), and myosin-II pulls the nodes together into a continuous contractile ring. At the end of anaphase a signaling system consisting of a GTPase and three protein kinases (the septation initiation network, SIN) triggers constriction of the contractile ring and associated synthesis of new cell wall to form a septum. The septum is a three-layered structure, with the primary septum flanked by two secondary septae. Digestion of the primary septum separates the daughter cells.
(Reference: Wu J-Q, Kuhn JR, Kovar DR, Pollard TD: Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev Cell 5:723–734, 2004.)
The assembly pathway is not yet understood in animal cells. INCENP and anillin move from the interphase nucleus to the cortex around the cell equator in early anaphase (Fig. 44-23). Some preexisting actin filaments are recruited intact into the contractile ring from adjacent areas of the cortex, whereas other filaments form de novo in the developing ring by assembly from monomers. Formins and profilin are involved, so the pathway resembles that in fission yeast. Myosin-II for the contractile ring is derived from various interphase structures. In cultured vertebrate cells, most of the myosin-II comes from stress fibers (see Fig. 33-1) that break down during prophase. Myosin-II is dispersed throughout the cytoplasm until anaphase, when it concentrates in the cortex, especially around the equator where the furrow forms.
Constriction of the Cleavage Furrow
Cleavage furrow ingression is widely believed to be driven by contraction of the contractile ring, though some researchers believe that alterations in the physical characteristics (e.g., tension and stiffness) of the plasma membrane may also have a role to play. Constriction of the ring probably involves a sliding filament mechanism, similar to muscle (see Figs. 39-12 and 39-20). During the early stages of furrowing, the contractile ring maintains a constant volume, but the structure disassembles completely by the end of cleavage. Thus, during the later stages of cytokinesis, contraction is accompanied by disassembly of the ring.
Constriction of the contractile ring is regulated so that it does not begin until after the onset of anaphase B, when sister chromatids are well separated. Local release of calcium appears to initiate constriction of the contractile ring in some cells. The calcium may activate the enzyme myosin light chain kinase, which, in turn, activates myosin-II (see Fig. 39-21). Exposure of dividing cells to agents that stimulate the release of calcium accelerates the appearance and rate of propagation of the cleavage furrow, whereas injection of compounds that bind calcium can inhibit cytokinesis. Other kinases and counterbalancing phosphatases are also involved, so regulation of myosin-II in cytokinesis is actually quite complex.
Membrane Addition and Abscission
As the contractile ring pulls the cell membrane inward, the single cell that entered mitosis is gradually transformed into two daughter cells joined by a thin intercellular bridge (Fig. 44-20). This process requires a significant net increase in the surface area of the cell. New plasma membrane is inserted adjacent to the leading edge of the furrow. The source of the new membrane appears to be secretory vesicles derived from the Golgi apparatus, and addition to the cleavage furrow is a specialized form of exocytosis. Fusion of vesicles providing the new membrane depends on specific syntaxins, t-SNAREs (see Chapter 21) that promote vesicle fusion along the secretory pathway. Targeted endocytosis is also important for cytokinesis, although its role is unclear.
In most animal cells, contraction of the cleavage furrow ultimately reduces the cytoplasm to a thin intercellular bridge between the two daughter cells. The intercellular bridge contains a highly ordered, antiparallel array of microtubules derived from the spindle with a dense knob, the midbody, at its center (Fig. 44-20). Isolated midbodies contain over 160 proteins, about one third involved in various aspects of membrane trafficking.
The midbody is encircled by a dense ring of proteins that includes the kinesin-6 that is essential for central spindle assembly (see the earlier section titled “Assembly and Regulation of the Contractile Ring”) and a protein known as centriolin, which is associated with the centrosome for the rest of the cell cycle. A related yeast protein regulates the exit from mitosis. The conserved domain of centriolin binds the exocyst, a multisubunit protein complex that targets secretory vesicles to the plasma membrane (see Chapter 21, under the section titled “Tethering Factors”). These secretory vesicles accumulate near the midbody and fuse with the plasma membrane to separate the two daughter cells from one another. The details of this fusion event are not yet understood. The exocyst complex also contributes to cytokinesis in budding and fission yeasts.
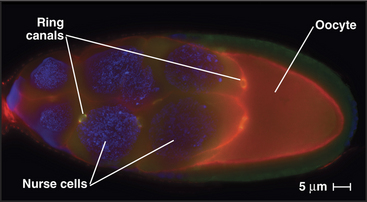
In some tissues, intercellular bridges remain open as ring canals. After several rounds of nuclear division with incomplete cytokinesis, the network of cells maintains cytoplasmic continuity as each former contractile ring matures into a larger ring canal. During Drosophila oogenesis, four rounds of nuclear division with persistent ring canals creates 15 nurse cells, all in continuity with the oocytes (Fig. 44-25). The cytoplasmic continuity through ring canals allows nurse cells to transfer their cytoplasm into the developing egg, thus greatly increasing its stockpile of proteins and mRNAs available for use in early development. In mammals, incomplete cytokinesis is notable in the testis, where ring canals connect several hundred developing sperm cells.
Exit from Mitosis
In budding yeast, a signaling pathway called the mitotic exit network (MEN) terminates mitosis, promotes contraction of the contractile ring, and initiates septation. The pathway consists of a small GTPase and protein kinases. Cdk kinase activity suppresses the pathway until anaphase, when Cdk activity drops sharply. The MEN GTPase is associated with one spindle pole body (the yeast version of the centrosome), while its key regulator, a GTP exchange factor, is located in the bud. Elongation of the mitotic spindle during anaphase B moves the GTPase into the bud, where it is activated.
Albertson R, Riggs B, Sullivan W. Membrane traffic: A driving force in cytokinesis. Trends Cell Biol. 2005;15:92-101.
Balasubramanian MK, Bi E, Glotzer M. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr Biol. 2004;14:R806-R818.
Burgess DR, Chang F. Site selection for the cleavage furrow at cytokinesis. Trends Cell Biol. 2005;15:156-162.
Collas P, Courvalin J-C. Sorting nuclear membrane proteins at mitosis. Trends Cell Biol. 2000;10:5-8.
Hirano T. Chromosome cohesion, condensation, and separation. Annu Rev Biochem. 2000;69:115-144.
Jürgens G. Plant cytokinesis: Fission by fusion. Trends Cell Biol. 2005;15:277-283.
Mitchison TJ, Salmon ED. Mitosis: A history of division. Nat Cell Biol. 2001;3:E17-E21.
Nasmyth K, Peters JM, Uhlmann F. Splitting the chromosome: Cutting the ties that bind sister chromatids. Science. 2000;288:1379-1385.
Piekny A, Werner M, Glozer M. Cytokinesis: Welcome to the Rho zone. Trends Cell Biol. 2005;15:651-658.
Rappaport R. Cytokinesis in Animal Cells: Developmental and Cell Biology Series. Cambridge, England: Cambridge University Press, 1996.
Sharp DJ, Rogers GC, Scholey JM. Microtubule motors in mitosis. Nature. 2000;407:41-47.
Sullivan SM, Maddock JR. Bacterial division: Finding the dividing line. Curr Biol. 2000;10:R249-R252.
Vagnarelli P, Earnshaw WC. Chromosomal passengers: The four dimensional regulation of mitotic events. Chromosoma. 2004;113:211-222.
Von Dassow G, Bement WM. A ring-like template for abscission. Dev Cell. 2005;9:578-580.
Wittmann T, Hyman A, Desai A. The spindle: A dynamic assembly of microtubules and motors. Nat Cell Biol. 2001;3:E28-E34.