Erysipelothrix, Lactobacillus, and Similar Organisms
1. Describe the Gram stain morphology of Arcanobacterium, Lactobacillus, Erysipelothrix, and Gardnerella spp.
2. Identify the media of choice and morphologic appearance of Gardnerella sp. and describe its incubation conditions, including time, oxygen requirements, and temperature.
3. List the disease states associated with Erysipelothrix, Gardnerella, and Lactobacillus spp.
4. Identify the correct specimens for the isolation of Erysipelothrix, Gardnerella, and Lactobacillus spp.
5. Explain why in vitro susceptibility testing is usually not necessary to guide therapy of Erysipelothrix or Gardnerella spp.
General Characteristics
The genera described in this chapter are all catalase-negative, non-spore-forming, gram-positive rods; some may exhibit rudimentary branching. Erysipelothrix rhusiopathiae is one of three species in the genus, but it is considered the only human pathogen. E. rhusiopathiae consists of several serovars based on peptidoglycan structure. The serovars most commonly associated with human infection include serovars 1 and 2. Arcanobacterium spp. demonstrate irregular, gram-positive rods on Gram stain. Gardnerella sp. fermentation byproducts include acetic and lactic acid. The cell wall of Gardnerella sp. is significantly thinner and contains less peptidoglycan than the typically gram-positive bacteria. Weissella confusa, formerly classified as Lactobacillus confusus, is included in Tables 18-3 and 18-4 because it is easily confused on culture media with the organisms included in this chapter, and in rare cases it has been isolated associated with bacteremia and endocarditis.
Epidemiology
Erysipelothrix spp. are found worldwide in a variety of vertebrate and invertebrate animals, including mammals, birds, and fish. Other domestic animals that may be infected include sheep, rabbits, cattle, and turkeys. The organism may be transmitted through direct contact or ingestion of contaminated water or meat. Arcanobacterium spp. are normal inhabitants of the mucosal membranes of cattle, sheep, dogs, cats, and pigs. The organisms listed in Table 18-1 include those that are closely associated with animals and are contracted by humans through animal exposure (e.g., E. rhusiopathiae and Arcanobacterium pyogenes) and those that are part of the normal human flora (e.g., Lactobacillus spp. and Gardnerella vaginalis).
TABLE 18-1
| Species | Habitat (Reservoir) | Mode of Transmission |
| Erysipelothrix rhusiopathiae | Normal flora; carried by and causes disease in animals | Zoonoses; abrasion or puncture wound of skin with animal exposure |
| Arcanobacterium haemolyticum | Normal flora of human skin and pharynx | Uncertain; infections probably caused by person’s endogenous strains |
| Arcanobacterium pyogenes | Normal flora; carried by and causes disease in animals. | Uncertain: Abrasion or undetected wound during exposure to animals |
| Gardnerella vaginalis | Normal flora: Human vaginal tissue Colonizers: Distal urethra of males |
Endogenous strain |
| Lactobacillus spp. | Environmental: Widely distributed in foods and nature Normal flora: Human mouth, gastrointestinal tract, and female genital tract |
Endogenous strain Infections are rare. |
Pathogenesis and Spectrum of Disease
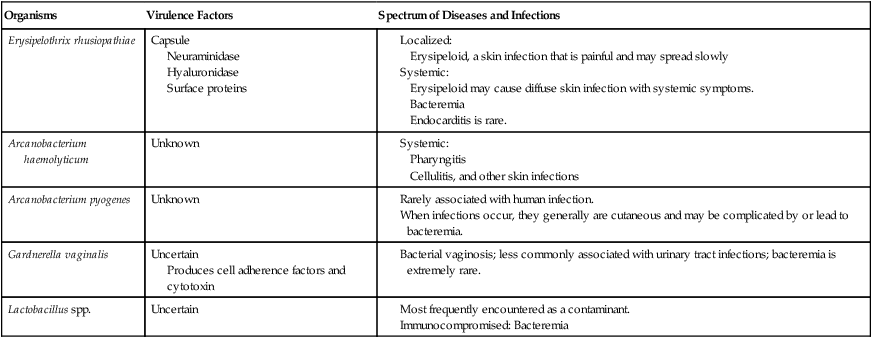
G. vaginalis and Lactobacillus spp. (Table 18-2) are natural inhabitants of the human vagina. Vaginal infections with G. vaginalis are often found in association with a variety of mixed anaerobic flora. Extravaginal infections are uncommon but have been identified associated with postpartum endometritis, septic abortion, and cesarean birth.
TABLE 18-2
Pathogenesis and Spectrum of Disease
| Organisms | Virulence Factors | Spectrum of Diseases and Infections |
| Erysipelothrix rhusiopathiae | Capsule Neuraminidase Hyaluronidase Surface proteins |
Produces cell adherence factors and cytotoxin
Laboratory Diagnosis
Specimen Collection and Transport
Generally, no special considerations are required for specimen collection and transport of the organisms discussed in this chapter. Of note, skin lesions for Erysipelothrix should be collected by biopsy of the full thickness of skin at the leading edge of the discolored area. Refer to Table 5-1 for other general information on specimen collection and transport.
Direct Detection Methods
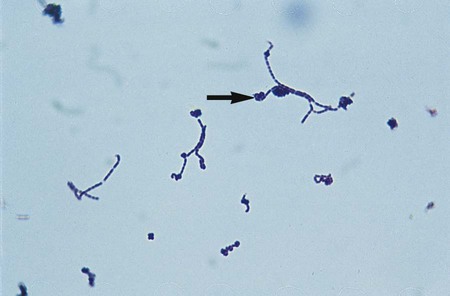
Lactobacillus is highly pleomorphic, occurring in long chaining rods and in coccobacilli and spiral forms (Figure 18-1).
Colonial Appearance
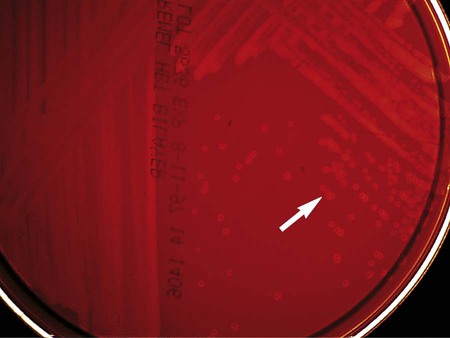
Table 18-3 describes the colonial appearance and other distinguishing characteristics (e.g., hemolysis) of each genus on sheep blood agar. G. vaginalis produces small, gray, opaque colonies surrounded by a diffuse zone of beta-hemolysis on HBT agar (Figure 18-2).
TABLE 18-3
Colonial Appearance on 5% Sheep Blood Agar and Other Characteristics
| Organism | Appearance |
| Arcanobacterium spp. | Small to large colonies with various appearances, including smooth, mucoid, and white and dry, friable, and gray; may be surrounded by narrow zone of beta-hemolysis |
| Erysipelothrix rhusiopathiae | Two colony types: large and rough or small, smooth, and translucent; shows alpha-hemolysis after prolonged incubation |
| Gardnerella vaginalis | Pinpoint; nonhemolytic |
| Lactobacillus spp. | Multiple colonial morphologies, ranging from pinpoint, alpha-hemolytic colonies resembling streptococci to rough, gray colonies |
| Weissella confusa | Pinpoint; alpha-hemolytic and may be confused with organisms presented in this chapter. |
Approach to Identification
The identification of the four genera described in this chapter must be considered along with that of Actinomyces, Bifidobacterium, and Propionibacterium spp., which are discussed in Chapter 42. Although the latter genera are usually considered with the anaerobic bacteria, they grow on routine laboratory media in 5% to 10% CO2. Some are catalase negative. Therefore, as shown in Table 18-4, these organisms must be considered together when a laboratory encounters catalase-negative, gram-positive, non-spore-forming rods.
TABLE 18-4
| Urease | Nitrate Reduction | Beta-Hemolysisb | FERMENTATIONa OF: | Other Comments | |||||||
| Glucose | Maltose | Mannitol | Sucrose | Xylose | CAMPc | GLCd | |||||
| Actinomyces israelii | − | v | − | + | + | v | + | + | − | A, L, S | |
| A. odontolyticus | − | + | −e | + | v | − | + | v | − | A, S | Red pigment produced after 1 week on SBA |
| A. naeslundii | + | + | − | + | + | v | + | v | − | A, L, S | |
| A. radingae | − | − | −w | + | + | − | + | + | − | S? | Pyrazinamidase, beta-galactosidase−positive and esculin-positive |
| A. turicensis | − | − | −w | + | v | − | + | + | − | NT | Pyrazinamidase, beta-galactosidase−negative and esculin-negative |
| A. graevenitzii | − | − | − | + | + | − | + | − | ND | L > S | |
| Actinobaculum schaalii | − | − | − | + | + | − | v | + | +w | A, s | Beta-galactosidase−negative |
| Arcanobacterium haemolyticum | − | − | + | + | + | − | v | − | Reverse +f | A, L, S | Gelatin-negative at 48 hr; beta-hemolysis is stronger on agar containing human or rabbit blood |
| A. pyogenes | − | − | +g | + | v | v | v | + | − | A, L, S | Gelatin-positive at 48 hr; casein-positive |
| A. bernardiae | − | − | − | + | + | − | − | − | − | A, L, S | |
| Bifidobacterium adolescentis | − | − | − | + | + | − | + | + | ND | A > L (s) | |
| Erysipelothrix sp. | − | − | − | +h | − | − | − | − | A, L, S | H2S-positive in TSI butt; vancomycin-resistant; alpha-hemolytic | |
| Lactobacillus spp. | − | − | − | + | + | v | + | ND | ND | L (a s) | Some strains vancomycin-resistant; alpha-hemolytic |
| Propionibacterium acnes | − | + | − | + | − | − | − | − | + | A, P (iv L s) | Indole-positive; may show beta-hemolysis on rabbit blood agar |
| P. propionicumj | − | + | − | + | + | + | + | − | ND | A, P, S, (L) | Colony may show red fluorescence under long-wavelength UV light |
| Gardnerella vaginalis | − | − | − | + | + | − | v | −k | ND | A (l s) | Beta-hemolysis on HBT; usually hydrolyses hippurate |
| Weissella spp. | NT | − | − | + | + | − | + | v | NT | L (as) | Vancomycin-resistant, small, short rods; produces gas from MRS broth; alpha-hemolytic; esculin-positive; arginine-positive |
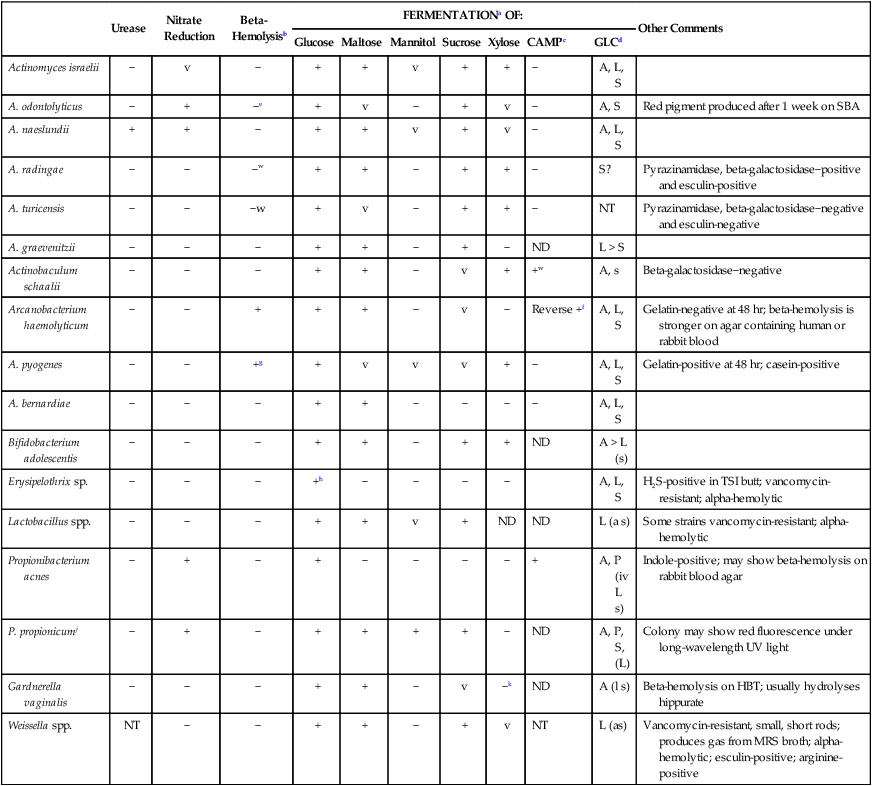
HBT, Human blood bilayer Tween agar; iv, isovaleric acid; ND; not done, NT; not tested; SBA, 5% sheep blood agar; TSI, triple sugar iron agar; v, variable; w, weak; +, ≥90% of strains positive; −, ≥90% of strains negative.
iSome strains are catalase negative.
aFermentation is detected in peptone base with Andrade’s indicator.
cCAMP test using a beta-lysin–producing strain of Staphylococcus aureus.
dEnd products of glucose metabolism: A, Acetic acid; L, lactic acid; P, propionic acid; S, succinic acid; ( ), may or may not produce acid end product.
eMay show beta-hemolysis on brain-heart infusion agar with sheep or human blood.
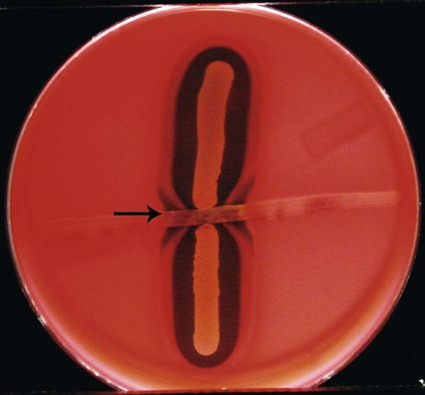
fReverse CAMP test; Staphylococcus aureus beta-lysins are inhibited by a diffusible substance produced by A. haemolyticum (Figure 18-3).
gMay also show beta-hemolysis on brain-heart infusion agar with human blood.
hReaction may be weak or delayed.
Comments Regarding Specific Organisms
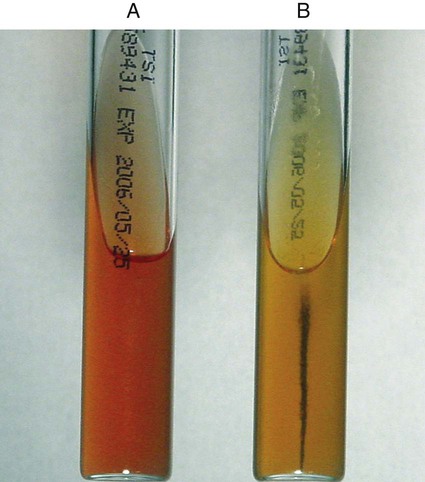
Erysipelothrix sp. is the only catalase-negative, gram-positive non-spore-forming rod that produces hydrogen sulfide (H2S) when inoculated into triple sugar iron (TSI) agar (Figure 18-4). Some Bacillus spp. also blacken the butt of TSI, but they are catalase positive and produce spores. Automated identification with the Vitek2 and Phoenix systems and the API ID system is reliable for identification.
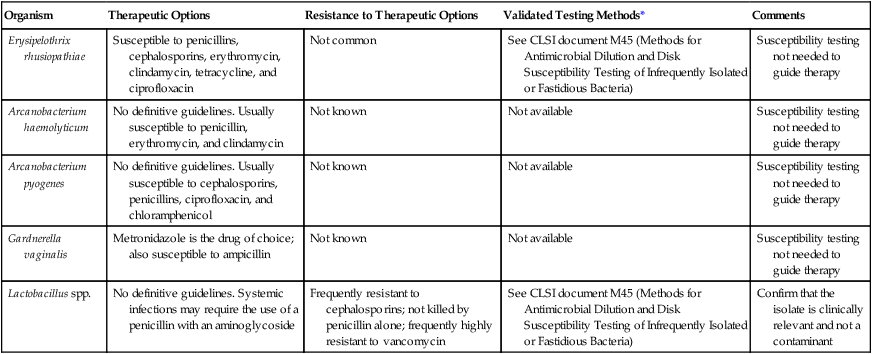
Antimicrobial Susceptibility Testing and Therapy
The rarity with which most of these organisms are encountered as the cause of infection has made the development of validated in vitro susceptibility testing methods difficult (Table 18-5). However, most of the organisms are susceptible to the agents used to eradicate them, therefore in vitro testing is not usually necessary to guide therapy. Lactobacillus spp. can be resistant to various antimicrobial agents. Fortunately, these organisms are rarely implicated in infections. When they are encountered in specimens from normally sterile sites, careful evaluation of their clinical significance is warranted before any attempt is made at performing a nonstandardized susceptibility test.
TABLE 18-5
Antimicrobial Therapy and Susceptibility Testing
| Organism | Therapeutic Options | Resistance to Therapeutic Options | Validated Testing Methods* | Comments |
| Erysipelothrix rhusiopathiae | Susceptible to penicillins, cephalosporins, erythromycin, clindamycin, tetracycline, and ciprofloxacin | Not common | See CLSI document M45 (Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria) | Susceptibility testing not needed to guide therapy |
| Arcanobacterium haemolyticum | No definitive guidelines. Usually susceptible to penicillin, erythromycin, and clindamycin | Not known | Not available | Susceptibility testing not needed to guide therapy |
| Arcanobacterium pyogenes | No definitive guidelines. Usually susceptible to cephalosporins, penicillins, ciprofloxacin, and chloramphenicol | Not known | Not available | Susceptibility testing not needed to guide therapy |
| Gardnerella vaginalis | Metronidazole is the drug of choice; also susceptible to ampicillin | Not known | Not available | Susceptibility testing not needed to guide therapy |
| Lactobacillus spp. | No definitive guidelines. Systemic infections may require the use of a penicillin with an aminoglycoside | Frequently resistant to cephalosporins; not killed by penicillin alone; frequently highly resistant to vancomycin | See CLSI document M45 (Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria) | Confirm that the isolate is clinically relevant and not a contaminant |
*Validated testing methods include standard methods recommended by the Clinical and Laboratory Standards Institute (CLSI) and commercial methods approved by the U.S. Food and Drug Administration (FDA).
Although some of these organisms may grow on the media and under the conditions recommended for testing other bacteria (see Chapter 12 for more information regarding validated testing methods), this does not necessarily mean that interpretable and reliable results will be produced. Chapter 12 should be reviewed for preferable strategies that can be used to provide susceptibility information when validated testing methods do not exist for a clinically important bacterial isolate.