CHAPTER 12 Chromosome Organization
Chromosomes are enormous DNA molecules that can be propagated stably through countless generations of dividing cells (Fig. 12-1). Genes are the reason for the existence of the chromosomes, but in higher eukaryotes, they actually make up only a small fraction of the chromosomal DNA, much of which does not encode proteins or other known functional RNAs. Cells package chromosomal DNA with roughly twice its weight of protein. This DNA-protein complex, called chromatin, is discussed in Chapter 13.
(From Paulson JR, Laemmli UK: The structure of histone-depleted chromosomes. Cell 12:817–828, 1977.)
In addition to the genes, only three classes of specialized DNA sequences are needed to make a fully functional chromosome: (1) a centromere, (2) two telomeres, and (3) an origin of DNA replication for approximately every 100,000 base pairs (bp). Centromeres regulate the partitioning of chromosomes during mitosis and meiosis. Telomeres protect the ends of the chromosomal DNA molecules and ensure their complete replication. DNA replication is discussed in Chapter 42. Chapter 15 considers the structure of genes. Box 12-1 lists a number of key terms presented in this chapter.
BOX 12-1 Key Terms
Chromosome Morphology and Nomenclature
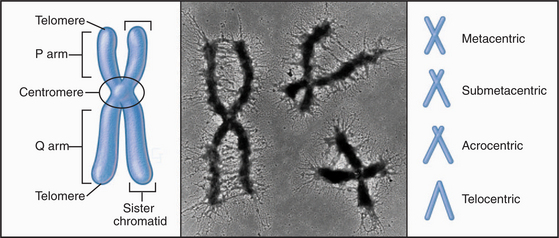
With few specialized exceptions, chromosomes from somatic cells of higher eukaryotes are visualized directly only during mitosis. Each mitotic chromosome consists of two sister chromatids that are held together at a waist-like constriction called the centromere. The portions of the chromosomes that are not in the centromere itself are called chromosome “arms” (Fig. 12-2).
One DNA Molecule per Chromosome
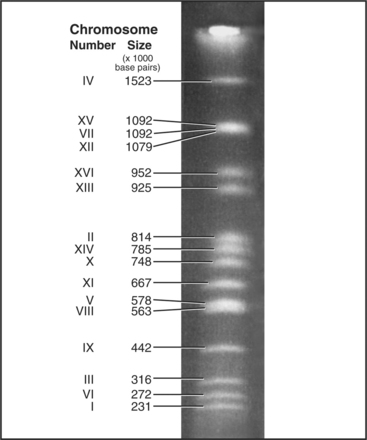
Each eukaryotic chromosome contains one DNA molecule that stretches between the telomeres at either end. Most prokaryotic and mitochondrial chromosomes are circular DNA molecules that lack telomeres, but naturally occurring eukaryotic nuclear chromosomes are generally linear DNA molecules with two telomeres. The clearest proof that each chromosome is composed of a single DNA molecule has been obtained for budding and fission yeasts, where intact chromosomal DNA molecules may be visualized by pulsed-field gel electrophoresis as a characteristic series of bands (Fig. 12-3). This technique can display the largest chromosome of fission yeast at 5,598,923 bp, but even the smallest human chromosome, which is about 40 million bp long, is too large to resolve in this way.
The Organization of Genes on Chromosomes
The first chromosome to be completely sequenced (in 1977) was that of the bacterial virus φx174 (Table 12-1). Starting in the 1990s much effort worldwide has been devoted to determining the complete sequences of the chromosomes of a wide variety of organisms (see Fig. 2-4). Sequencing efforts that have been completed to date have generated an enormous bank of data on the genetic composition of simple and complex organisms. For example, over 100 microbial genomes have been sequenced. One major goal of this effort—the sequence of the human genome—is now essentially complete.
| Organism | Haploid Genome Size (bp) | Predicted Number of Protein-Coding Genes |
|---|---|---|
| φX174 (bacterial virus) | 5386 | 11 |
| Mycoplasma genitalium (pathogenic bacterium) | 580,070 | 480* |
| Rickettsia prowazekii (endoparasitic bacterium) | 1,111,523 | 834 |
| Escherichia coli (free-living bacterium) | 4,639,221 | 4288 |
| Bacillus subtilis (free-living bacterium) | 4,214,810 | 4100 |
| Saccharomyces cerevisiae (budding yeast) | 14,000,000 | 6604 |
| Schizosaccharomyces pombe (fission yeast) | 13,800,000 | 4824 |
| Caenorhabditis elegans (nematode worm) | 9.7 × 107 | 19,100 |
| Drosophila melanogaster (fruit fly) | 1.4 × 108 | 13,525 |
| Arabadopsis thaliana (plant) | 1.25 × 108 | 25,498 |
| Anopheles gambiae (malaria mosquito) | 2.78 × 108 | 14,000 |
| Oryza sativa japonica (rice) | 4.2 × 108 | 32,000–50,000 |
| Mus musculus (house mouse) | 2.6 × 109 | −30,000 |
| Rattus norvegicus (Brown Norway rat) | 2.75 × 109 | −21,000–46,000 |
| Xenopus laevis (South African clawed frog) | 3.1 × 109 | ? |
| Homo sapiens (human) | 3.1 × 109 | 20,000–25,000 |
| Triturus cristatus (salamander) | 2.2 × 1010 | ? |
Note: In most higher eukaryotes, with the exception of some plants, the huge tracts of repeated DNA sequences in and around centromeres are poor in genes and beyond the limits of present technology to sequence. Thus, when statistics are given on chromosome sizes in descriptions of genome sequencing projects, these portions are generally omitted. Where possible, the genome size figures given here reflect the entire genome (sequenced and unsequenced).
* It appears that only 265 to 350 of these genes are essential for life.
Complex genomes that have been sequenced thus far range in size from 580,000 bp for Mycoplasma genitalium, which causes urinary tract infections in humans to 2,863,476,365 bp for humans themselves. Numbers of protein-coding genes identified range from 480 in M. genitalium to 20,000 to 25,000 for humans (Table 12-1). However, because gene prediction algorithms are still being perfected, only rough estimates of gene number are available, even for completely sequenced genomes.
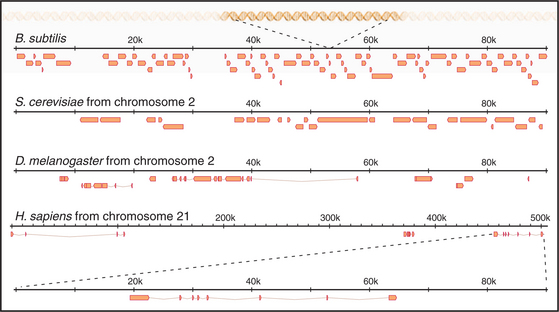
The first eukaryote whose genome was entirely sequenced was the budding yeast Saccharomyces cerevisiae. The 14 million bp yeast genome is subdivided into 16 chromosomes ranging in size from 230,000 bp to over 1 million bp (Fig. 12-3). This genome has a dramatic history. Ancestral budding yeast apparently had eight chromosomes but at one point underwent a duplication of the entire genome. This event was followed by numerous small deletions that resulted in the subsequent loss of most of the duplicated genes, with about 10% remaining. As a result, the modern budding yeast genome contains about 5700 predicted genes, many of which are paralogs (genes produced by duplication that have evolved to take on distinct functions; see Box 2-1). As a result, only about 1000 of these genes are indispensable for life. About 5% of yeast genes are segmented, containing regions that appear in mature RNA molecules (exons) and regions that are removed by splicing (introns) (discussed in detail in Chapter 16). Exons occupy approximately 75% of the budding yeast genome, with the remainder in regulatory regions, repeated DNAs, and introns (Fig. 12-4).
The next genome sequences to be completed were those of two very important “model” organisms that have been widely used by cell and developmental biologists: the nematode worm Caenorhabditis ele-gans and the fruit fly Drosophila melanogaster. These sequences revealed a number of important organizational differences from budding yeast. Although its ge-nome is eight times larger than that of budding yeast (97 million bp distributed in six chromosomes), the nematode has only about three times more genes. Surprisingly, the fly, despite its even larger genome and more complex body plan and life cycle, has about one third fewer genes than the worm. In fact, only about 27% of the C. elegans genome and 13% of the Drosophila genomic DNA code for proteins. Instead, the fly has much more noncoding repetitive DNA than the worm.
The “finished” sequence of the human genome, published in 2004, revealed an even lower density of genes. Humans have far fewer genes than had been predicted: about 20,000 to 25,000, in contrast to some earlier predictions of up to 100,000 (Table 12-1). Protein-coding regions occupy only about 1.2% of the chromosomes. In contrast, various repeated-sequence elements and pseudogenes appear to occupy about 50% of the genome, as is discussed in a later section. To put this all in perspective, every million bp of DNA sequenced yielded 483 genes in S. cerevisiae, 197 genes in C. elegans, 117 genes in D. melanogaster, and only 7 to 9 genes in humans. If the Escherichia coli chromosome were the size of chromosome 21, the smallest human chromosome at ˜40 × 106 bp, it would have nearly 37,000 genes—more than the entire human complement! In fact, chromosome 21 is predicted to have only 225 genes.
Transposons Make Up Much of the Human Genome
Even though humans no longer have active transposons, we still use at least two functional vestiges of these elements. It has been known for years that one of the ways in which the diversity of the immune system is generated is by cutting and pasting portions of the genes that encode the variable regions of the immunoglobulin chains (see Fig. 28-10). This process involves moving bits of DNA around, and it now appears that the enzymes that accomplish this process were originally encoded by ancient transposons. In addition, CENP-B (centromere protein B; see Fig. 13-23), an abundant protein that binds to the α-satellite DNA repeats in primate centromeres, is closely related to a transposase enzyme encoded by one family of transposons.
The best-known retrotransposons are LINES (long interspersed nuclear elements) and SINES (short interspersed nuclear elements). Reverse transcriptases encoded by LINES are responsible for movements of both LINES and SINES. The L1 class of LINES encodes two proteins, one of which has reverse transcriptase activity (Fig. 12-5). All DNA polymerases, including reverse transcriptases, work by elongating a preexisting stretch of double-stranded nucleic acid (see Chapter 42 for a discussion of the mechanism of DNA synthesis). L1 elements insert themselves into the chromosome by first nicking the chromosomal DNA, then using the newly created end as a primer for synthesis of a new DNA strand (Fig. 12-5). The template for this DNA synthesis by the reverse transcriptase is the LINE RNA, and the newly synthesized DNA is made as a direct extension of the chromosomal DNA molecule. Most LINES are only partial copies of the full-length element. Apparently, the reverse transcriptase is not very efficient (processive): It usually falls off before it completes copying the entire element.
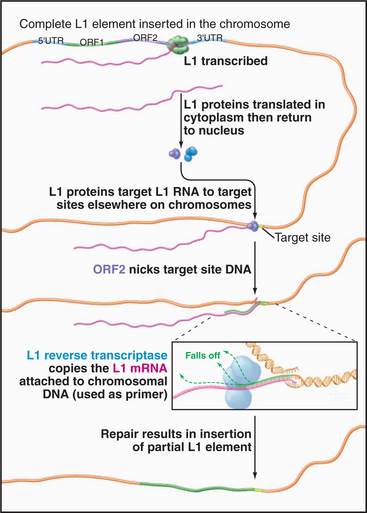
Figure 12-5 mechanism of transposition of an l1 element. The element is transcribed by RNA polymerase II (see Fig. 15-4). Proteins encoded by the element nick the chromosome, promote base pairing of the L1 transcript with the target site, and reverse transcribe the RNA into DNA. The L1 DNA is synthesized as an extension of the chromosome. The mechanism of final closing up of the nicks and gaps is not yet fully understood.
LINES and SINES plus other remnants of transposable elements account for up to 45% of the human genome. LINES, with a consensus sequence of 6 to 8 kb, make up about 20% of the genome. (A consensus sequence is the average arrived at by comparing a number of different sequenced DNA clones.) About 79% of human genes have at least one segment of L1 sequence inserted, typically in an intron. The Alu class of SINES, with a consensus sequence of about 300 bp, constitutes about 13% of the total DNA—almost a million copies scattered throughout the genome. Alu elements are derived from the 7SL RNA gene, which encodes the RNA component of signal recognition particle (see Fig. 20-5). They are actively transcribed by RNA polymerase III (see Fig. 15-10) but are short and do not have enough coding capacity to encode for complex proteins. They therefore rely on the L1 machinery to move around. It is therefore somewhat paradoxical that SINES and LINES have quite different distributions along the chromosomes. LINES are concentrated in gene-poor regions of the chromosomes with a relatively higher content of A + T base pairs. In contrast, the Alu SINES are concentrated in gene-rich regions with a relatively higher content of G + C base pairs.
Transposition can be harmful, as along the way, genes can be disrupted, deleted, or rearranged. Because of their tendency to insert into gene-rich regions of chromosomes, Alu elements are one of the most potent endogenous human mutagens, with a new Alu insertion occurring once in every 100 births. In contrast, although LINES can cause genome instability when they move, several factors appear to minimize the damage that they cause. Despite the large fraction of the human genome that is derived from LINES, they cause only 0.07% of spontaneous mutations seen in humans, owing to several mitigating features: (1) Only about 60 to 100 L1 elements are active, and these appear to be active only in the germ line (i.e., during production of gametes). (2) LINE elements prefer to move into gene-poor areas of chromosomes. (3) Most LINE sequences are only fragments of the complete element. In contrast, mice apparently have many more active L1 elements (˜3000), and L1 transposition causes about 2.5% of spontaneous mutations in mice. One of the ancestral roles of the RNAi machinery (see Fig. 16-12) might have been to suppress the deleterious activity of transposable elements.
The physiological role, if any, of these elements is much debated. One long-favored possibility is that they do nothing advantageous and are analogous to an infection of the DNA that is tolerated as long as it does not move into genes that are essential for life. This is called the “selfish DNA” hypothesis. This notion has been challenged for Alu sequences, which are efficiently transcribed into RNA. Alu transcripts accumulate under conditions of cellular stress such as viral infection. This is interesting, because Alu transcripts can bind very efficiently to a protein kinase called PKR, which is induced by interferon as part of the cell’s antiviral protection pathways. The best-known function of PKR is phosphorylation of eukaryotic initiation factor 2a-subunit (eIF-2a; see Fig. 17-9). This profoundly inhibits protein synthesis. PKR is generally activated by dsRNAs, and this is presumably important for its antiviral role, as many viruses have RNA chromosomes. Alu transcripts at low levels activate PKR (i.e., suppress protein synthesis), but at higher levels, they inactivate the enzyme (i.e., promote protein synthesis). Thus, it has been suggested that Alu transcripts might be natural regulators of protein synthesis under conditions of cellular stress.
Pseudogenes
One surprise that emerged from analysis of the human genome sequence was the number of pseudogenes, which may exceed the number of genes. Pseudogenes are derived from genes but are no longer functional. They arise in two ways, both involving transposable elements. Processed pseudogenes, the more common variety, are created by reverse transcription of mature mRNA sequences into DNA apparently by a LINE re-verse transcriptase that then inserts the copy back into the genome. Because these sequences come from ma-ture mRNA, they lack introns. They also lack sequences that regulate transcription initiation and termination (Chapter 15), so they are not expressed. Unprocessed pseudogenes are created either by reverse transcription of unspliced precursor mRNAs or by local duplications of the chromosome that generally occur as a result of recombination between transposable elements. Such duplications initially create bona fide functional gene copies that become pseudogenes as they accumulate mutations that render their transcripts nonfunctional. Because pseudogenes are not functional, mutation of their DNA is not selected against during evolution, as are harmful mutations in the coding sequences of genes. Thus, over time, pseudogenes become less and less recognizable, and they eventually are lost from recognition in the sea of noncoding DNA.
Segmental Duplications in the Human Genome
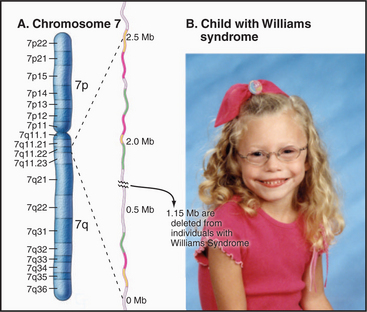
One example of this is found on chromosome 7, where deletion of a portion of the long arm is associated with Williams-Beuren syndrome, a complex developmental disorder associated with a highly variable range of symptoms that can include elfin-like facial features, defects in certain mental skills, and a wide range of physical problems (Fig. 12-6). These deletions, which typically remove about 1.6 × 106 bp, occur because of large segmental duplications of blocks of >140,000 bp distributed across a region of 2 × 106 bp. These duplications flank a unique sequence region of 1.15 × 106 bp that is lost when recombination occurs between the regions of segmental duplication. Because of the highly complex organization of this region and the large size of the duplications, this turned out to be the most difficult region of chromosome 7 to sequence, and in fact, some ambiguities still remain.
The Centromere: Overview
The centromere is at the heart of all chromosomal movements in mitosis and meiosis, as it is the region where the chromosome becomes attached to the mitotic spindle (the microtubule-based apparatus upon which chromosomes move; see Chapter 44). The centromere also has an important role in monitoring the attachment of the chromosomes to the spindle and controlling the progress of cells through mitosis. The centromere is a nucleoprotein structure, and both DNA and proteins are essential to its function.
In chromosomes of most higher eukaryotes, the centromere may be visualized directly as a waist-like stricture or primary constriction where the two sister chromatids are most intimately paired. Several abundant families of repetitive DNAs are concentrated in the centromeres of human chromosomes. The chromatin of human centromeres is entirely embedded within constitutive heterochromatin, a form of chromatin characterized by the presence of special proteins and special modifications of the histone proteins, that is generally nonpermissive for gene transcription and that remains condensed throughout the cell cycle (see Fig. 13-9). At least some of the repeated DNA elements are transcribed into dsRNA, and the cellular RNAi machinery is required for the assembly of heterochromatin. (For a discussion of heterochromatin, see Chapter 13.) At the surface of the centromeric heterochromatin is a button-like structure called the kinetochore, which directs chromosomal movements in mitosis (see Fig. 13-20). Kinetochores are packaged into a specialized form of chromatin called centrochromatin.
Variations in Centromere Organization among Species
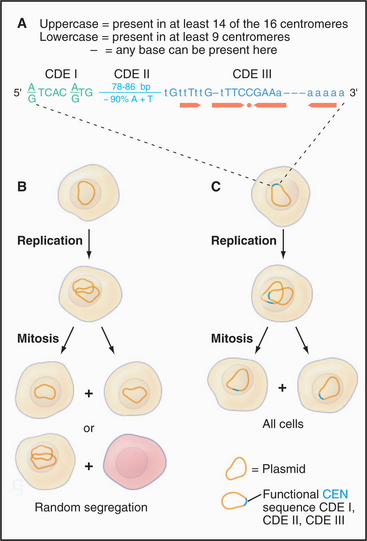
Each chromosome has particular DNA sequences, called CEN sequences, that specify protein-binding sites required for assembly of the kinetochore. In budding yeast, CEN sequences are autonomous; if inserted into circular DNA molecules (plasmids), they can render them capable of interacting with the mitotic spindle and segregating during mitosis (Fig. 12-7). In other organisms, including the fission yeast Schizosaccharomyces pombe, CEN sequences appear to require an activation event in order to nucleate kinetochore formation. This event appears to involve some sort of modification of the DNA and/or chromatin (discussed later).
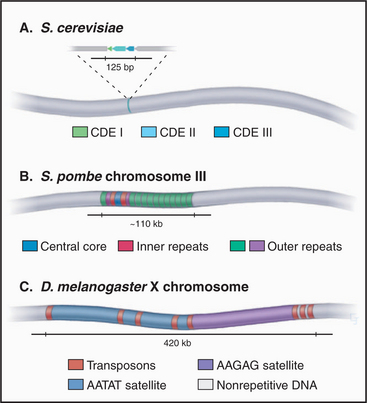
CEN sequences from all 16 chromosomes of budding yeast have a common organization based around three conserved sequence elements (Fig. 12-7). These are designated (in the 5′ to 3′ direction) CDE I (centromere DNA element I, 8 bp), CDE II (78 to 86 bp), and CDE III (25 bp). A 125-bp region spanning CDE I to CDE III is sufficient to direct the efficient segregation of a yeast chromosome, which can reach a size of more than 1 million bp. This type of centromere, in which the kinetochore is assembled as a result of protein recognition of specific DNA sequences, is known as a point centromere. Kinetochores assembled on point centromeres bind a single microtubule.
Even though the average size of S. pombe chromosomes is only fivefold larger than their counterparts in S. cerevisiae (4.6 mb versus 0.87 mb), fission yeast centromeres are 300- to 600-fold larger (Fig. 12-8). The smallest S. pombe centromere consists of 35,000 bp, whereas the largest spans 110,000 bp. Fission yeast centromeres are also much more complex than are their counterparts from budding yeast, containing a central core of unique-sequence DNA that is 4 to 7 kb long, flanked by complex arrays of repeated sequences. This type of centromere, where the kinetochore is assembled on a variable array of repeated DNA sequences, is known as a regional centromere. Kinetochores assembled on regional centromeres bind multiple microtubules (2 to 4 in the case of S. pombe).
In both S. pombe and metazoans, these epigenetic changes involve the construction of a special chromatin environment. S. pombe kinetochores assemble into a spe-cialized form of chromatin containing the kinetochore-specific histone variant CENP-A (see Fig. 13-23). This flanking heterochromatin is important because it is required for binding a protein complex that regulates the pairing of replicated chromosomal DNA molecules during mitosis. Mutations in S. pombe that impair formation of heterochromatin also impair centromere function.
The best-characterized centromere of a metazoan comes from rice, in which the centromere of chromosome 8 has been completely sequenced. This was possible because this particular centromere contains relatively limited amounts of the rice centromeric satellite DNA (CentO), and this is dispersed in a number of blocks separated by transposons, retrotransposons, and fragments. All in all, 72% of this centromere is composed of repetitive sequences. The kinetochore, as defined by sequences associated with the kinetochore-specific histone H3 variant CENP-A (see Fig. 13-23), spans 750 kb and appears to be interspersed with regions of chromatin containing normal histone H3. The state of posttranslational modifications of that H3 indicates that it is packaged heterochromatin (a “closed” state of the chromatin usually associated with the absence of active gene transcription; see Fig. 13-9). It was therefore surprising that this centromere region contains at least four genes that are actively transcribed. The structure of this centromere appears to be intermediate between that of a canonical vertebrate centromere and a neocentromere (see later); however, rice centromere 8 is not a new variant but has had this organization for at least the last 10,000 years, since the indica and japonica cultivars of rice were separated.
The centromere organization of the fruit fly D. melanogaster shows important similarities and differences to the plant centromeres. The centromere of the fly’s X chromosome is contained within a stretch of roughly 420,000 bp (Fig. 12-8) that is composed mostly of simple-sequence satellite DNAs interspersed with transposable DNA elements. This resembles the situation in plants; however, in Drosophila, no sequences were found in this region that are unique to the fly centromeres; all sequences that are found at centromeres can also be found on the chromosome arms. Thus, it appears that something other than the DNA sequence alone must be responsible for conferring centromere activity on this region of the chromosome.
Most higher eukaryotes have regional centromeres, but a third variant is also observed in many insects as well as in the nematode C. elegans. Holocentric chromosomes have centromere activity distributed along the whole surface of the chromosome during mitosis. Thus, instead of having a tight bundle of 20 microtubules binding to a disk-like kinetochore at a centromeric constriction, as in humans, in C. elegans about 20 microtubules bind at scattered sites along the whole poleward-facing surface of the chromosome during mitosis. If a holocentric chromosome is fragmented, all the pieces have the ability to bind microtubules and segregate in mitosis. Perhaps surprisingly, the proteins of the holocentric kinetochore are the same as those found at disk-like regional kinetochores (see Chapter 13). At the moment, nothing is known about the DNA sequences, presumably interspersed throughout the genome, that direct the assembly of holocentric kinetochores. One possibility is that in these chromosomes, any chromatin can serve to nucleate kinetochore assembly—perhaps the requirement for special epigenetic marks has been relaxed.
Mammalian Centromere DNA
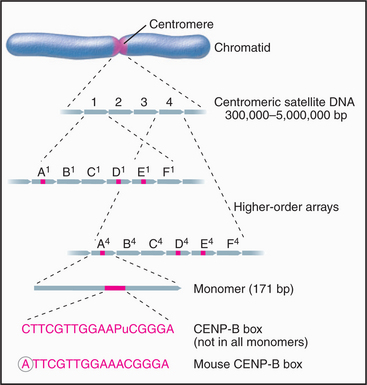
The major human centromeric satellite DNA, α-satellite, is a complex family of repeated sequences that constitutes approximately 5% of the genome. Monomers averaging around 171 bp long are organized into higher-order repeats (Fig. 12-9). Some of the monomers have a conserved 17-bp sequence (the CENP-B box), which forms the binding site for the centromeric protein CENP-B (mentioned earlier as having its origin in an ancient DNA transposon). The organization of higher-order repeats varies greatly from chromosome to chromosome, and numerous repeat patterns, comprising 2 to 32 monomers, have been described. Each chromosome has one or a few types of higher-order repeats of α-satellite DNA.
What is human CEN DNA, that is, the DNA that nucleates kinetochore formation? The best candidate thus far is α-satellite, which occurs at all natural centromeres. The entire centromeric region of certain chromosomes may be composed of α-satellite monomers, apparently with little or no interspersed DNA of other types. The amount of α-satellite DNA at different centromeres varies widely: from as little as 300,000 bp on the Y chromosome to up to 5 million bp on chromosome 7. In addition, the α-satellite DNA content of a given chromosome can vary by more than a million bp between different individuals. Thus, whatever the function of α-satellite DNA may be, clearly, a wide variation in local organization is tolerated.
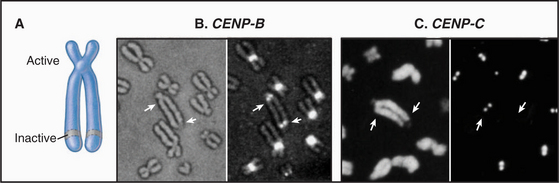
The epigenetic mark that defines an active centromere can be lost as well as gained. Thus, it is possible for a centromere to retain its normal DNA composition and yet lose the ability to assemble a kinetochore. This has been seen most clearly in naturally occurring human dicentric chromosomes. The example shown in Figure 12-10 arose through a breakage and fusion near the long arm of chromosome 13. Thus, it has two centromeres. As shown in the figure, one of these, even though it retains its α-satellite, has lost the ability to assemble a kinetochore.
Once a DNA sequence has acquired the proper epigenetic mark, it can assemble a functional kinetochore that can regulate chromosome behavior in mitosis. This involves the binding and function of a great many proteins, to be discussed in Chapter 13 (see Fig. 13-23).
The Ends of the Chromosomes: Why Specialized Telomeres Are Needed
The ends of chromosomal DNA molecules pose at least two problems that cells solve by packaging the chromosome ends into specialized structures called telomeres. First, it is essential that cells distinguish the ends of a chromosome from breaks in DNA. When cells detect DNA breaks, they stop their progression through the cell cycle and repair the breaks by joining the ends together (see Box 43-1). Telomeres keep normal chromosome ends from inducing cell cycle arrest and from being joined to other DNA ends by the repair machinery. Second, telomeres permit the chromosomal DNA to be replicated out to the very end (see later dis-cussion).
How Telomeres Replicate the Ends of the Chromosomal DNA
One role of telomeres is to prevent the erosion of the end of the chromosomal DNA molecule during each round of replication (for a more extensive discussion of DNA replication, see Chapter 42). All DNA replication proceeds with a polarity of 3′ to 5′ on the template DNA (5′ to 3′ in the newly synthesized DNA). Furthermore, all DNA polymerases (but not RNA polymerases) work by elongating a preexisting stretch of double-stranded nucleic acid. During cellular DNA replication, this is achieved by making a short RNA primer and then elongating the RNA: DNA duplex with DNA polymerase. The primer is subsequently removed, and the newly opened gap is filled by a DNA polymerase elongating from the next upstream DNA end (Fig. 12-11).
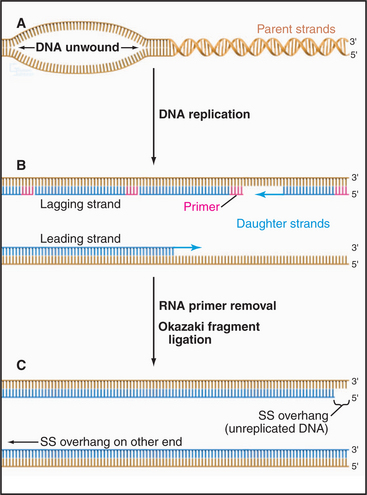
Figure 12-11 the dna replication problem at chromosome ends. DNA polymerases cannot initiate the formation of DNA on a template de novo; they can only extend preexisting nucleotide strands (see Chapter 42). In contrast, RNA polymerases can initiate synthesis without a primer. All replicating DNA chains start from a short region of RNA, which is used to “prime” DNA polymerase. A, DNA strand separation. B, RNA primer synthesis. Replication of DNA starts with the synthesis of an RNA primer complementary to a short sequence of DNA, which is extended by DNA polymerase. C, The RNA primer is degraded and the gap is filled in by DNA polymerase. This being true, how is the DNA underneath the very last RNA primer replicated?
If the terminus of the chromosomal DNA is replicated from an RNA primer that sits on the very end of the DNA molecule, it follows that when this primer is removed, there is no upstream DNA on which to put a primer. How, then, is the DNA underneath the last RNA primer replicated? Years of searching for a DNA polymerase that could operate in the opposite direction proved fruitless. The answer that ultimately emerged turned out to be both elegant and unexpected.
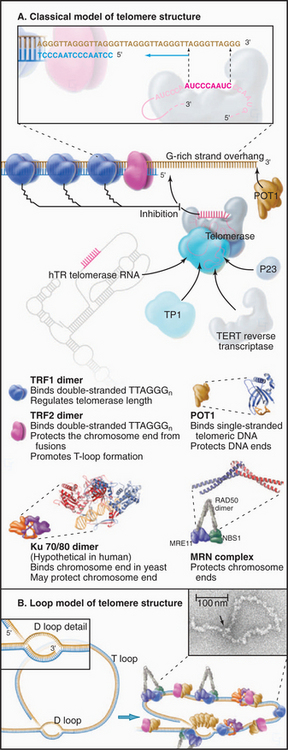
The sequence of the human telomere is TTAGGG. When the RNA component of human telomerase was characterized, it was found to contain the sequence AUCCCAAUC, which could base-pair with the TTAGGG repeat at the end of the chromosome. This observation led to a proposal for the mechanism of telomerase action (Fig. 12-12). In brief, the enzyme uses its own RNA as a template for the synthesis of DNA, which it attaches to the end of the chromosome. This hypothesis has been confirmed by studies that show that alterations in the sequence of the telomerase RNA lead to alterations in the telomere sequence at the end of the chromosome.
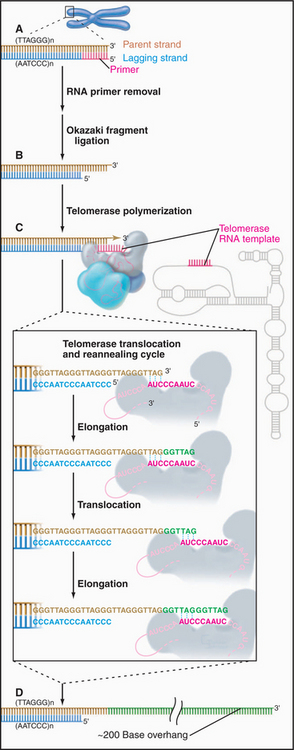
Figure 12-12 telomerase provides a special mechanism for lengthening chromosomal ends. a–b, Normal mechanisms of DNA replication are unable to replicate the very 3′ end of the chromosomal DNA. C, Telomerase solves this problem by providing its own template in the form of an intrinsic RNA subunit. This RNA subunit contains a sequence complementary to that found at the chromosome terminus on the 3′ strand. This sequence is able to base-pair with the DNA at the chromosome terminus and act as a template for DNA synthesis. In this case, the primer is the 3′ end of the chromosomal DNA, and the template is the RNA of the telomerase enzyme. Thus, the process of telomere elongation is a specialized form of reverse transcription (copying RNA into DNA), a process similar to that occurring during transposition of LINE elements (see Fig. 12-5), and most well known during the life cycle of certain RNA-containing tumor viruses. The telomerase enzyme releases and rebinds its template after each 6 to 7 bp of new DNA has been synthesized. Up to several hundred bp may be added to the telomere in this way. D, In most cells, the cycle of telomerase activity and subsequent DNA replication leaves a single-stranded G-rich strand about 200 nucleotides long.
According to this model, the telomerase actually synthesizes DNA from an RNA template. Thus, telomerase is a reverse transcriptase similar to that involved in the movement of the LINE retrotransposons (see Fig. 12-5). Interestingly, the L1 family of LINE retrotransposons appear to insert themselves into the chromosome by a similar mechanism, in which a DNA end created at a nick in the chromosome is used to prime synthesis of a DNA strand using the LINE RNA as template, the newly synthesized DNA being a direct extension of the chromosomal DNA molecule.
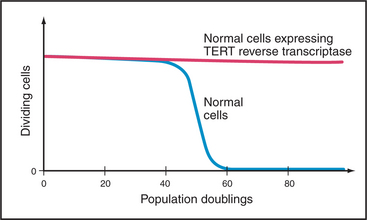
Telomerase is subject to tight biological regulation, active enzyme being detected in only a few normal tissues of adult humans. These include the intestine and testis. In addition, about 85% of cancer cells express active telomerase. In cells that lack telomerase, an alternative pathway, thought to involve DNA recombination, can help to maintain the telomeric repeats at chromosome ends. Paradoxically, hTR and TP1 are not tightly regulated. They are detected in many tissues, most of which lack telomerase activity. By contrast, the expression of hTERT correlates tightly with telomerase activity. Furthermore, introduction of a DNA-encoding hTERT into telomerase-negative cells is sufficient to convert those cells to telomerase-positive. This can have extremely important consequences for the proliferation of the cells (Fig. 12-15).
Structural Proteins of the Telomere
Telomeres provide special protected ends for the chromosomal DNA molecule, in part by coating the end of the DNA molecules with protective proteins and by adopting a specialized DNA loop structure. In organisms in which telomeric DNA sequences are relatively short, these sequences are packaged into a specialized chromatin structure. In mammals, in which the telomeric sequences are much longer, the bulk of the telomeric DNA is packaged into conventional chromatin (see Chapter 13).
Three types of proteins associate with telomeres (Fig. 12-13). The first binds in a sequence-specific manner to the double-stranded telomeric repeats. In budding yeast, the best known such protein is Rap1p. Mammals have two essential proteins of this type: TRF1 and TRF2 (telomere repeat factor). TRF1 and its associated factors regulate telomerase activity, thus helping to maintain the proper length of telomeres. A similar function has been proposed for Rap1p in budding yeast, where the telomeres appear to elongate only until they create a threshold number of binding sites for this protein. TRF2 and its associated factors protect the chromosome ends; interference with the binding of this protein to telomeres results in a loss of the G-strand overhangs and a dramatic increase in the tendency of chromosomes to fuse end to end. This may be because TRF2 can promote the formation of a special looped configuration of DNA in which the single-stranded G-strand overhang is base-paired with “upstream” DNA (Fig. 12-13B). Figure 12-14 shows the phenotype of a Drosophila mutant that lacks a protein essential for the assembly of the proper protective structure at telomeres and therefore undergoes extensive chromosome fusion.
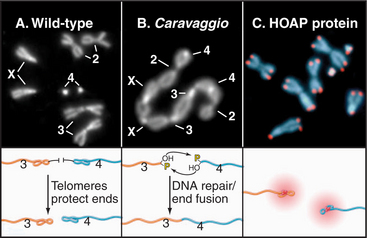
Figure 12-14 disruption of the protective complex at telomeres results in chromosome fusion. A, The chromosomes of a wild-type female Drosophila melanogaster seen at mitotic metaphase (see Chapter 44). B, The caravaggio mutant is characterized by a “train” of chromosomes generated by telomere-telomere fusions. (Caravaggio is the name of an Italian train.) C, The cav gene encodes HP1/Orc2 Associated Protein (HOAP), which specifically localizes at all Drosophila telomeres.
(Images courtesy of Gianni Cenci and Maurizio Gatti, University of Rome, Italy. B, From Cenci G, Siriaco G, Raffa GD, et al: Drosophila HOAP protein is required for telomere capping. Nat Cell Biol 5:82–84, 2003. C, Part of montage on Nature Cell Biology cover.)
Telomeres, Aging, and Cancer
The relationship between telomere length and aging can be studied in cultured cells. Normal cells in culture grow for only a limited number of generations (often called the Hayflick limit) before undergoing senescence (this involves cessation of growth, enlargement in size, and expression of marker enzymes, such as β-galactosidase). Because normal somatic cells lack telomerase activity, their telomeres shorten by about 50% before the cells senesce. Senescent cells stop dividing before their telomeres become critically short. In some cases, it is possible to force the senescent cells to resume proliferation (e.g., by expressing certain viral oncogenes). These “driven” cells continue to divide and their telomeres continue to shorten until a crisis point is reached. In crisis, cells suffer chromosomal instability (chromosomal fusions and breaks can occur) and cell death. In populations of human cells in crisis, very rarely (in about 1 in a million cases), cells appear that once again grow normally. These cells now express telomerase. These observations with cultured cells led to the suggestion that senescence might occur in cells when the telomeric repeats of one or more chromosomes are reduced to some critical level.
In a second experiment, the hTERT reverse transcriptase subunit of telomerase was introduced into normal cells growing in culture. This caused an increase in the level of active telomerase with dramatic results. Instead of undergoing senescence, these cells kept dividing in culture, apparently indefinitely (Fig. 12-15). However, unlike cancer cells, which are also immortal, these cells did not acquire the ability to cause tumors. Thus, this experiment showed convincingly that telomeres are part of a mechanism that regulates the proliferative capacity of somatic cells.
de Lange T. T-loops and the origin of telomeres. Nat Rev Mol Cell Biol. 2004;5:323-329.
Doolittle RF. Microbial genomes opened up. Nature. 1998;392:339-342.
Dujon B. The yeast genome project: What did we learn? Trends Genet. 1996;12:263-270.
Fukagawa T. Centromere DNA, proteins and kinetochore assembly in vertebrate cells. Chromosome Res. 2004;12:557-567.
Hall AE, Keith KC, Hall SE, et al. The rapidly evolving field of plant centromeres. Curr Opin Plant Biol. 2004;7:108-114.
International Human Genome Sequencing Consortiuim. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931-945.
Kim SH, Kaminker P, Campisi J. Telomeres, aging and cancer: In search of a happy ending. Oncogene. 2002;21:503-511.
Maiato H, DeLuca J, Salmon ED, Earnshaw WC. The dynamic kinetochore-microtubule interface. J Cell Sci. 2004;117:5461-5477.
McAinsh AD, Tytell JD, Sorger PK. Structure, function, and regulation of budding yeast kinetochores. Annu Rev Cell Dev Biol. 2003;19:519-539.
McEachern MJ, Krauskopf A, Blackburn EH. Telomeres and their control. Annu Rev Genet. 2000;34:331-358.
Pidoux AL, Allshire RC. Centromeres: Getting a grip of chromosomes. Curr Opin Cell Biol. 2000;12:308-319.
Rubin GM, Yandell MD, Wartman JR, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204-2215.
Shay JW, Zou Y, Hiyama E, Wright WE. Telomerase and cancer. Hum Mol Genet. 2001;10:677-685.
Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 1999;9:657-663.
Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu Rev Biochem. 2005;73:177-208.