Viral Structure, Classification, and Replication
I Structure and Classification of Viruses
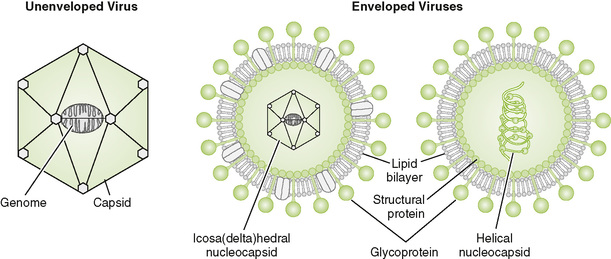
1. A virion, or viral particle, consists of a genome (DNA or RNA) packaged within a protein coat, the capsid, which may or may not be surrounded by a membrane envelope.
2. Essential enzymes or other proteins are carried within some viruses.
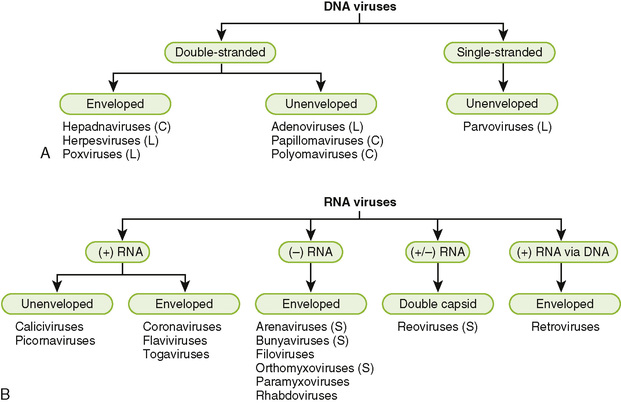
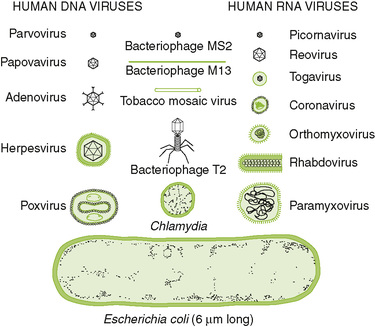
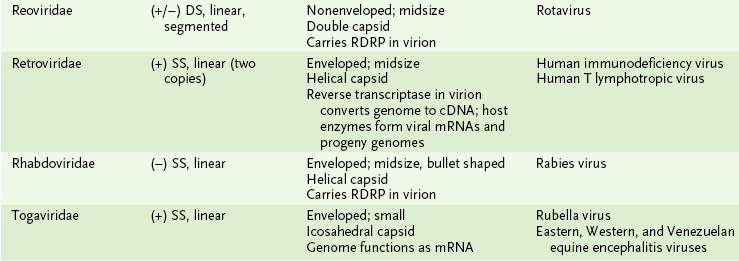
3. The major virus families can be classified based on their genome structure, size, and whether they are enveloped or not enveloped.
• Found in the reoviruses, a (+/−) RNA genome, and in three families of (−) RNA viruses (orthomyxoviruses, arenaviruses, and bunyaviruses)
• Consists of several pieces, or segments, each of which encodes at least one polypeptide
• May undergo reassortment among genomic segments, yielding new virus strains, particularly in influenza viruses
• In viruses that lack an outer envelope, the capsid enclosing the genome forms the outer layer of the virion.
• Icosahedral capsid is found in many simple viruses (e.g., picornaviruses); shape approximates a sphere with 12 vertices.
• Icosadeltahedral capsid is found in larger viruses (e.g., herpesviruses); shape is similar to a soccer ball.
• Helical capsid is found inside most viruses with (−) RNA genomes (e.g., paramyxoviruses).
3. Capsid components recognize and bind to cell surface receptors on host cells.
• Important differences between nonenveloped and enveloped viruses are summarized in Table 18-1.
TABLE 18-1
Nonenveloped (Naked) Versus Enveloped Viruses
| Property | Nonenveloped Viruses | Enveloped Viruses |
| Components | Proteins | Phospholipids, proteins, glycoproteins |
| Sensitivity to heat, acid, detergent, drying | Resistant (stable) | Sensitive (labile) |
| Release from host cell | By cell lysis (host cell killed) | By budding (host cell survives) and cell lysis |
| Transmission or mode of spread | Fomites, dust, fecal-oral | Large droplets, secretions, and organ or blood transplants |
| Effect of drying | Retain infectivity | Lose infectivity |
| Survival within gastrointestinal tract | Yes | No (except corona-and hepadna-viruses) |
| Host immune response (minimal protection) | Antibody response | Antibody and cell-mediated responses (the latter often contribute to pathogenesis) |
• Most enveloped viruses do not have a defined shape. Exceptions are the brick-shaped poxviruses and bullet-shaped rhabdoviruses.
• Viral envelopes are derived from host cell membranes into which viral structural proteins and glycoproteins are inserted (see Fig. 18-3).
3. Viral envelope glycoproteins that promote entry into target cells.
II Basic Steps in Viral Replication (Box 18-1; Fig. 18-5)
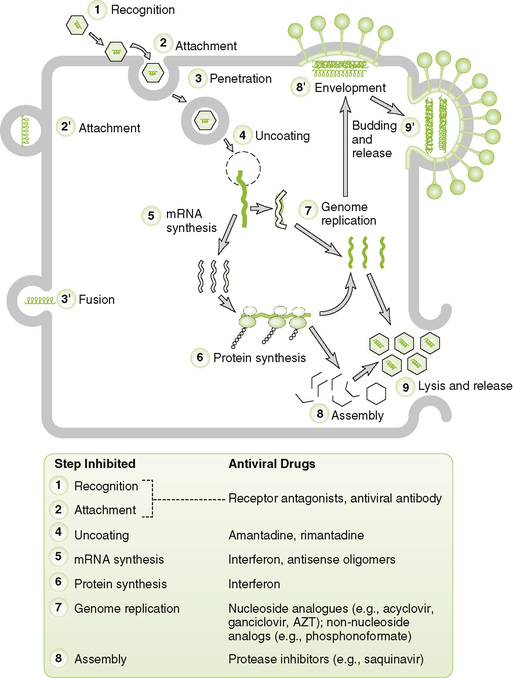
18-5 General scheme of virus replication. Enveloped viruses have alternative means of entry (steps 2’ and 3’), assembly (step 8’), and exit from the cell (step 9’). Antiviral drugs inhibit various steps, as indicated at the bottom. Antiviral drugs are described in Chapter 20 and for the relevant virus. mRNA, messenger RNA.
• Recognition step determines which cells will be infected (tropism or specificity of a virus) and is a major determinant of disease manifestations resulting from infection.
1. VAPs or other structures on the virion surface recognize tissue-specific receptors on target cells.
2. Cell surface virus receptors may be proteins, glycoproteins, or glycolipids. Examples of such receptors and the viruses that bind to them include the following:
• CD4 molecules on T cells and macrophages: HIV and human T lymphotropic virus
• CR2 receptor for C3b complement component on B cells and epithelial cells: Epstein-Barr virus
• Sialic acid side chains on membrane proteins or lipids: influenza viruses; paramyxoviruses (e.g., mumps, measles viruses)
• ICAM-1 on B lymphocytes, epithelial cells, and fibroblasts: rhinoviruses (common cold viruses)
C Entry (penetration) of virion into target cell
1. Receptor-mediated endocytosis: most nonenveloped and some enveloped viruses
• Endocytosed virions generally are released into cytoplasm as a result of decreased pH in endosomal vesicle or lysis of vesicle by virus.
2. Fusion of viral envelope with cell membrane
3. Viropexis (direct penetration of cell membrane by virions): reoviruses, picornaviruses
D Uncoating of nucleocapsid to release viral genome and enzymes
E Synthesis of viral mRNAs (Fig. 18-6)
1. Early mRNAs encode enzymes and control proteins required in small amounts (e.g., DNA-binding proteins).
2. Late mRNAs encode structural proteins required in large amounts (e.g., capsid proteins and glycoproteins).
3. DNA viruses depend on host cell machinery in the nucleus to make mRNAs from viral genomes.
• Exception is poxvirus, which uses RNA polymerase carried in virion to make mRNAs from viral genome in the cytoplasm.
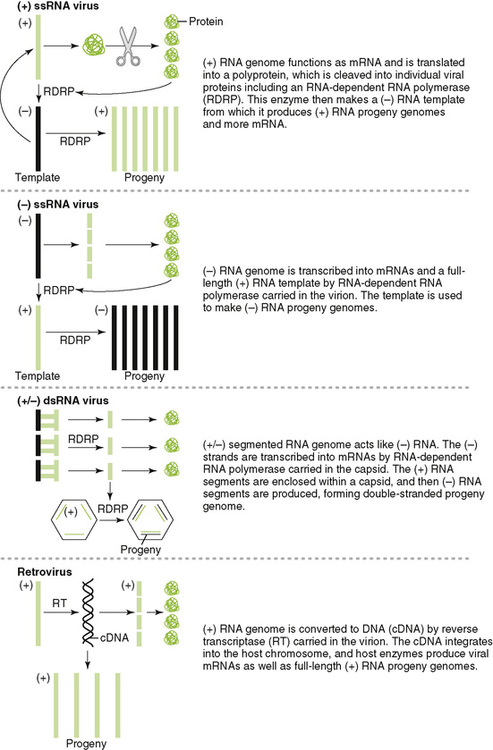
4. RNA viruses use several mechanisms for generating mRNA depending on the structure of the genome, as depicted in Figure 18-6.
• The RNA-dependent RNA polymerase is carried into the cell as part of the helical nucleocapsid by (−) RNA viruses and makes mRNA in the cytoplasm.
• The RNA-dependent RNA polymerase of (+) RNA viruses is made after infection of the cell and make a (−) RNA template and then copies new mRNA and new genomes from it.
1. Translation of mRNA into protein uses host cell ribosomes and other synthetic machinery.
2. Posttranslational modifications (e.g., glycosylation, phosphorylation, and proteolytic cleavage) are carried out by host enzymes and occasionally by viral enzymes.
1. DNA viruses replicate their genomes in the nucleus using host or virus-encoded DNA polymerases.
• Exceptions are poxvirus and hepadnavirus (hepatitis B virus), which replicate their genomes in the cytoplasm using viral enzymes.
2. RNA viruses (except retroviruses) use viral RNA-dependent RNA polymerase (replicase) to synthesize complementary (antisense) RNA, which acts as a template for synthesis of new genomes in the cytoplasm.
• Exception is orthomyxovirus (influenza virus), which is replicated in the nucleus but by viral enzymes.
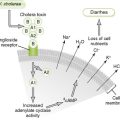
3. Retroviruses carry reverse transcriptase, a viral enzyme that converts (+) single-stranded RNA genome into double-stranded DNA, which is integrated into host chromosomal DNA.
• Transcription of integrated viral DNA (provirus) by host cell enzymes yields new retroviral genomes as well as mRNAs.
4. Hepadnaviruses use the cell’s DNA-dependent RNA polymerase to make an overlapping (+) RNA copy of the genome that is encapsidated with a reverse transcriptase that converts it into DNA.
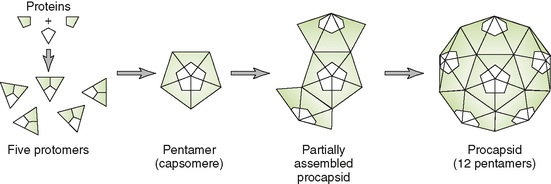
• Procapsid (empty shell) may assemble first and then be filled with the genome.
• Capsid proteins may assemble around the genome, forming the nucleocapsid.
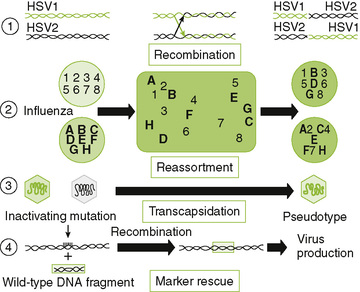
III Viral Genetic Mechanisms (Fig. 18-7)
• Antibody and antiviral drugs can select for resistant viruses that can be generated during infection of an individual.
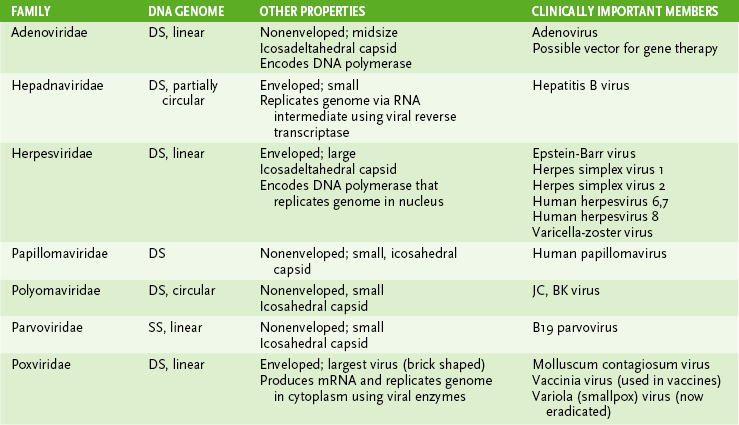
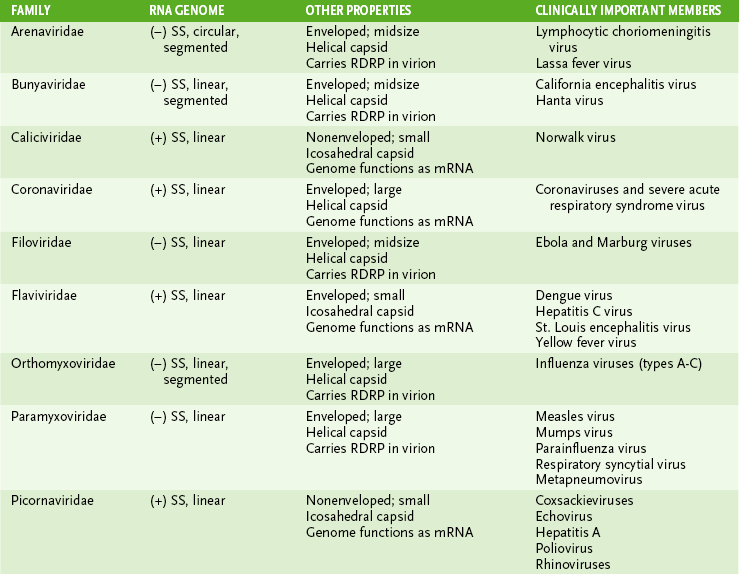
• Tables 18-2 and 18-3 summarize the structural properties of viral families and list important human pathogens in each.