Chapter 190 Salmonella
All Salmonella serovars form a single DNA hybridization group, a single species called S. enterica composed of several subspecies (Table 190-1). Each subspecies contains various serotypes defined by the O and H antigens. To further simplify the nomenclature for physicians and epidemiologists, the names for the common serovars are kept for subspecies I strains, which represent >99.5% of the Salmonella strains isolated from humans and other warm-blooded animals.
Table 190-1 SALMONELLA NOMENCLATURE
| TRADITIONAL USAGE | FORMAL NAME | CDC DESIGNATION |
|---|---|---|
| S. typhi | S. enterica* subsp. enterica ser. Typhi | S. ser. Typhi |
| S. dublin | S. enterica subsp. enterica ser. Dublin | S. ser. Dublin |
| S. typhimurium | S. enterica subsp. enterica ser. Typhimurium | S. ser. Typhimurium |
| S. choleraesuis | S. enterica subsp. enterica ser. Choleraesuis | S. ser. Choleraesuis |
| S. marina | S. enterica subsp. houtenae ser. Marina | S. ser. Marina |
CDC, U.S. Centers for Disease Control and Prevention; subsp, subspecies; ser., serovar.
* Some authorities prefer S. choleraesuis or S. enteritidis rather than S. enterica to describe the species.
190.1 Nontyphoidal Salmonellosis
Pathogenesis
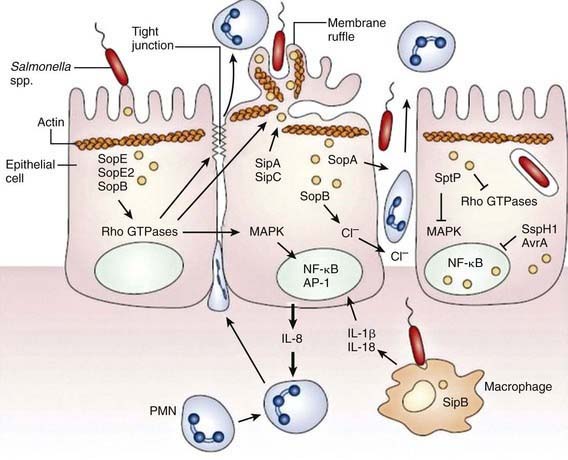
Although S. Typhimurium can cause systemic disease in humans, intestinal infection usually results in a localized enteritis that is associated with a secretory response in the intestinal epithelium. Intestinal infection also induces secretion of interleukin-8 (IL-8) from the basolateral surface and other chemoattractants from the apical surface, directing recruitment and transmigration of neutrophils into the gut lumen and thus preventing the systemic spread of the bacteria (Fig. 190-1).
Salmonella species invade epithelial cells in vitro by a process of bacteria-mediated endocytosis involving cytoskeletal rearrangement, disruption of the epithelial cell brush border, and the subsequent formation of membrane ruffles (Fig. 190-2). An adherent and invasive phenotype of S. enterica is activated under conditions similar to those found in the human small intestine (high osmolarity, low oxygen). The invasive phenotype is mediated in part by Salmonella pathogenicity island 1, a 40-kb region that encodes regulator proteins such as HilA, the type 3 secretory system involved in invasion of epithelial cells, and a variety of other products. In humans the TLR-dependent interleukin-12/interferon-λ (IL-12/IFN-λ) is a major immunoregulatory system that bridges innate and adaptive immunity and is responsible for restricting the systemic spread of nontyphoidal Salmonella.
Some virulence traits are shared by all salmonellae, but others are serotype restricted. These virulence traits have been defined in tissue culture and murine models, and it is likely that clinical features of human Salmonella infection will eventually be related to specific DNA sequences. With most diarrhea-associated nontyphoidal salmonelloses, the infection does not extend beyond the lamina propria and the local lymphatics. Specific virulence genes are related to the ability to cause bacteremia. These genes are found significantly more often in strains of S. Typhimurium isolated from the blood than in strains recovered from stool. Although both S. dublin and S. choleraesuis have a greater propensity to rapidly invade the bloodstream with little or no intestinal involvement, the development of disease after infection with Salmonella depends on the number of infecting organisms, their virulence traits, and several host defense factors. Various host factors may also affect the development of specific complications or clinical syndromes (Table 190-2) and of these, HIV infections are assuming greater importance in Africa in all age groups.
Table 190-2 HOST FACTORS AND CONDITIONS PREDISPOSING TO THE DEVELOPMENT OF SYSTEMIC DISEASE WITH NONTYPHOIDAL SALMONELLA STRAINS
Neonates and young infants (≤3 mo of age)
HIV/AIDS
Other immunodeficiencies and chronic granulomatous disease
Immunosuppressive and corticosteroid therapies
Malignancies, especially leukemia and lymphoma
Hemolytic anemia, including sickle cell disease, malaria, and bartonellosis
Collagen vascular disease
Inflammatory bowel disease
Achlorhydria or use of antacid medications
Impaired intestinal motility
Schistosomiasis, malaria
Malnutrition
Treatment
Appropriate therapy relates to the specific clinical presentation of Salmonella infection. In children with gastroenteritis, rapid clinical assessment, correction of dehydration and electrolyte disturbances, and supportive care, are key (Chapter 332). Antibiotics are not generally recommended for the treatment of isolated uncomplicated Salmonella gastroenteritis because they may suppress normal intestinal flora and prolong both the excretion of Salmonella and the remote risk for creating the chronic carrier state (usually in adults). However, given the risk for bacteremia in infants (<3 mo of age) and that of disseminated infection in high-risk groups with immune compromise (HIV, malignancies, immunosuppressive therapy, sickle cell anemia, immunodeficiency states), these children must receive an appropriate empirically chosen antibiotic until culture results are available (Table 190-3). The S. Typhimurium phage type DT104 strain is usually resistant to the following 5 drugs: ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline. An increasing proportion of S. Typhimurium phage type DT104 isolates also have reduced susceptibility to fluoroquinolones. Given the higher mortality associated with multidrug-resistant Salmonella infections, it is necessary to perform susceptibility tests on all human isolates. Infections with suspected drug-resistant Salmonella should be closely monitored and treated with appropriate antimicrobial therapy.
| ORGANISM AND INDICATION | DOSE AND DURATION OF TREATMENT |
|---|---|
| Salmonella infections in infants <3 mo of age or immunocompromised persons (in addition to appropriate treatment for underlying disorder) | Cefotaxime 100-200 mg/kg/day every 6 hr for 5-14 days |
| or | |
| Ceftriaxone 75 mg/kg/day once daily for 7 days | |
| or | |
| Ampicillin 100 mg/kg/day every 6 hr for 7 days | |
| or | |
| Cefixime 15 mg/kg/day for 7-10 days |
Prognosis
Most healthy children with Salmonella gastroenteritis recover fully. However, malnourished children and children who do not receive optimal supportive treatment (Chapters 55 and 332) are at risk for development of prolonged diarrhea and complications. Young infants and immunocompromised patients often have systemic involvement, a prolonged course, and extraintestinal foci. In particular, children with HIV infection and Salmonella infections can have a florid course.
After infection, nontyphoidal salmonellae are excreted in feces for a median of 5 wk.
Prevention
Control of the transmission of Salmonella infections to humans requires control of the infection in the animal reservoir, judicious use of antibiotics in dairy and livestock farming, prevention of contamination of foodstuffs prepared from animals, and use of appropriate standards in food processing in commercial and private kitchens (Table 190-4). Because large outbreaks are often related to mass food production, it should be recognized that contamination of just one piece of machinery used in food processing may cause an outbreak; meticulous cleaning of equipment is essential. Clean water supply and education in handwashing and food preparation and storage are critical to reducing person-to-person transmission. Salmonella may remain viable when cooking practices prevent food from reaching a temperature greater than 150°F (65.5°C) for >12 min. Parents should be advised of the risk of reptiles as pets in households with young infants.
Table 190-4 RECOMMENDATIONS FOR PREVENTING TRANSMISSION OF SALMONELLA FROM REPTILES AND AMPHIBIANS TO HUMANS
Pet store owners, health care providers, and veterinarians should provide information to owners and potential purchasers of reptiles and amphibians about the risks for and prevention of salmonellosis from these pets.
Persons at increased risk for infection or serious complications from salmonellosis (e.g., children aged <5 yr and immunocompromised persons) should avoid contact with reptiles and amphibians and any items that have been in contact with reptiles and amphibians.
Reptiles and amphibians should be kept out of households that include children aged <5 yr or immunocompromised persons. A family expecting a child should remove any pet reptile or amphibian from the home before the infant arrives.
Reptiles and amphibians should not be allowed in child-care centers.
Persons should always wash their hands thoroughly with soap and water after handling reptiles and amphibians or their cages.
Reptiles and amphibians should not be allowed to roam freely throughout a home or living area.
Pet reptiles and amphibians should be kept out of kitchens and other food preparation areas. Kitchen sinks should not be used to bathe reptiles and amphibians or to wash their dishes, cages, or aquariums. If bathtubs are used for these purposes, they should be cleaned thoroughly and disinfected with bleach.
Reptiles and amphibians in public settings (e.g., zoos and exhibits) should be kept from direct or indirect contact with patrons except in designated animal contact areas equipped with adequate handwashing facilities. Food and drink should not be allowed in animal contact areas.
From the Centers for Disease Control and Prevention: Reptile-associated salmonellosis—selected states, 1998-2002, MMWR Morbid Mortal Wkly Rep 52:1206–1210, 2003.
Centers for Disease Control and Prevention. Human salmonellosis associated with animal-derived pet treats—United States and Canada, 2005. MMWR Morbid Mortal Wkly Rep. 2006;55:702-705.
Centers for Disease Control and Prevention. Outbreak of multidrug-resistant Salmonella typhimurium associated with rodents purchased at retail pet stores—United States, December 2003-October 2004. MMWR Morbid Mortal Wkly Rep. 2005;54:429-434.
Centers for Disease Control and Prevention. Reptile-associated salmonellosis—selected states, 1998–2002. MMWR. 2003;52:1206-1210.
Chiu CH, Chuang CH, Chiu S, et al. Salmonella enterica serotype choleraesuis infections in pediatric patients. Pediatrics. 2006;117:e1193-e1196.
Coburn B, Grassl GA, Finlay BB. Salmonella, the host and disease: a brief review. Immunology Cell Biology. 2007;85:112-118.
Graham SM. Salmonellosis in children in developing and developed countries and populations. Curr Opin Infect Dis. 2000;15:507-512.
Hanning IB, Nutt JD, Ricke SC. Salmonellosis outbreaks in the United States due to fresh produce: sources and potential intervention measures. Foodborne Pathogens Dis. 2009;6:635-648.
Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nat Rev Microbiol. 2008;6:53-66.
Helms M, Simonsen J, Molbak K. Quinolone resistance is associated with increased risk of invasive illness or death during infection with Salmonella serotype Typhimurium. J Infect Dis. 2004;190:1652-1654.
Helms M, Vastrup P, Gerner-Smidt P, et al. Excess mortality associated with antimicrobial drug-resistant Salmonella typhimurium. Emerg Infect Dis. 2002;8:490-495.
Hohmann EL. Nontyphoidal salmonellosis. Clin Infect Dis. 2001;32:263-269.
International Food Safety Authorities Network. Antimicrobial-resistant Salmonella (pdf). www.who.int/foodsafety/fs_management/en/No_03_Salmonella_Apr05_en.pdf, 2010. Accessed September 16
Jones TF, Ingram LA, Fullerton KE, et al. A case-control study of the epidemiology of sporadic salmonella infection in infants. Pediatrics. 2006;118:2380-2387.
McEwen SA, Fedorka-Cray PJ. Antimicrobial use and resistance in animals. Clin Infect Dis. 2002;34(Suppl 3):S93-S106.
Morpeth SC, Ramadhani HO, Crump JA. Invasive non-Typhi Salmonella disease in Africa. Clin Infect Dis. 2009;49:606-611.
Santos RL, Tsolis RM, Baumler AJ, et al. Pathogenesis of Salmonella-induced enteritis. Braz J Med Biol Res. 2003;36:3-12.
Sirinavin S, Chiemchanya S, Vorachit M. Systemic nontyphoidal Salmonella infection in normal infants in Thailand. Pediatr Infect Dis J. 2001;20:581-587.
Stevens MP, Humphrey TJ, Maskell DJ. Molecular insights into farm animal and zoonotic Salmonella infections. Philos Trans R Soc Lond B Biol Sci. 2009;364:2709-2723.
Walsh AL, Phiri AJ, Graham SM, et al. Bacteremia in febrile Malawian children: clinical and microbiologic features. Pediatr Infect Dis J. 2000;19:312-318.
190.2 Enteric Fever (Typhoid Fever)
Pathogenesis
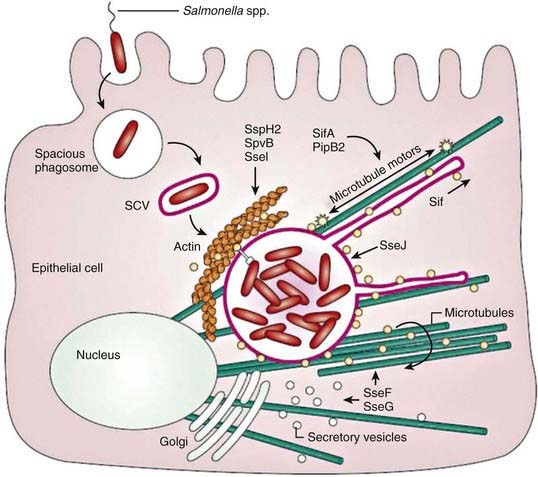
On contact with the epithelial cell, S. typhi assembles TTSS-1 and translocates effectors into the cytoplasm. These effectors activate host Rho guanosine triphosphatases (GTPases), resulting in the rearrangement of the actin cytoskeleton into membrane ruffles, induction of mitogen-activated protein kinase (MAPK) pathways, and destabilization of tight junctions. Changes in the actin cytoskeleton are further modulated by the actin-binding proteins SipA and SipC and lead to bacterial uptake. MAPK signaling activates the transcription factors activator protein-1 (AP-1) and nuclear factor-κB (NF-κB), which turn on production of IL-8. The destabilization of tight junctions allows the transmigration of polymorphonuclear leukocytes (PMNs) from the basolateral surface to the apical surface, paracellular fluid leakage, and access of bacteria to the basolateral surface. Shortly after internalization of S. Typhi by macropinocytosis, salmonellae are enclosed in a spacious phagosome that is formed by membrane ruffles. Later, the phagosome fuses with lysosomes, acidifies, and shrinks to become adherent around the bacterium, forming the Salmonella-containing vacuole (SCV). TTSS-2 is induced within the SCV and translocates effector proteins SifA and PipB2, which contribute to Salmonella-induced filament (Sif) formation along microtubules (see Fig. 190-2).
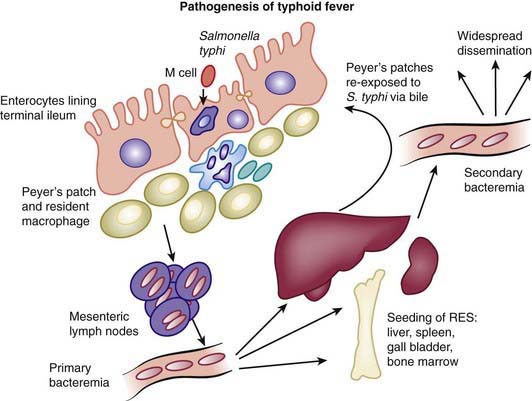
After passing through the intestinal mucosa, S. Typhi organisms enter the mesenteric lymphoid system and then pass into the bloodstream via the lymphatics. This primary bacteremia is usually asymptomatic, and blood culture results are frequently negative at this stage of the disease. The blood-borne bacteria are disseminated throughout the body and are thought to colonize the organs of the RES, where they may replicate within macrophages. After a period of bacterial replication, S. Typhi organisms are shed back into the blood, causing a secondary bacteremia that coincides with the onset of clinical symptoms and marks the end of the incubation period (Fig. 190-3).
Clinical Features
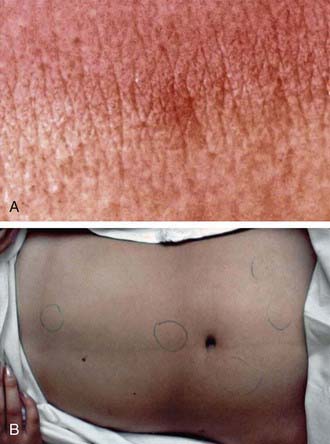
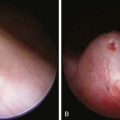
Typhoid fever usually manifests as high-grade fever with a wide variety of associated features, such as generalized myalgia, abdominal pain, hepatosplenomegaly, abdominal pain, and anorexia (Table 190-5). In children, diarrhea may occur in the earlier stages of the illness and may be followed by constipation. In the absence of localizing signs, the early stage of the disease may be difficult to differentiate from other endemic diseases such as malaria and dengue fever. The fever may rise gradually, but the classic stepladder rise of fever is relatively rare. In about 25% of cases, a macular or maculopapular rash (rose spots) may be visible around the 7th-10th day of the illness, and lesions may appear in crops of 10-15 on the lower chest and abdomen and last 2-3 days (Fig. 190-4). These lesions may be difficult to see in dark-skinned children. Patients managed as outpatients present with fever (99%) but have less emesis, diarrhea, hepatomegaly, splenomegaly, and myalgias than patients who require admission to the hospital.
Table 190-5 COMMON CLINICAL FEATURES OF TYPHOID FEVER IN CHILDREN*
| FEATURE | RATE (%) |
|---|---|
| High-grade fever | 95 |
| Coated tongue | 76 |
| Anorexia | 70 |
| Vomiting | 39 |
| Hepatomegaly | 37 |
| Diarrhea | 36 |
| Toxicity | 29 |
| Abdominal pain | 21 |
| Pallor | 20 |
| Splenomegaly | 17 |
| Constipation | 7 |
| Headache | 4 |
| Jaundice | 2 |
| Obtundation | 2 |
| Ileus | 1 |
| Intestinal perforation | 0.5 |
Complications
Rare complications include toxic myocarditis, which may manifest as arrhythmias, sinoatrial block, or cardiogenic shock (Table 190-6). Neurologic complications are also relatively uncommon among children; they include delirium, psychosis, increased intracranial pressure, acute cerebellar ataxia, chorea, deafness, and Guillain-Barré syndrome. Although case fatality rates may be higher with neurologic manifestations, recovery usually occurs with no sequelae. Other reported complications include fatal bone marrow necrosis, DIC, hemolytic-uremic syndrome, pyelonephritis, nephrotic syndrome, meningitis, endocarditis, parotitis, orchitis, and suppurative lymphadenitis.
Treatment
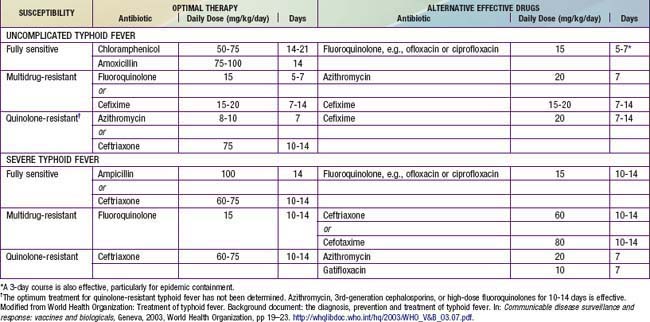
There are general principles of management of typhoid. Adequate rest, hydration, and attention are important to correct fluid and electrolyte imbalance. Antipyretic therapy (acetaminophen 10-15 mg/kg every 4-6 hr by mouth [PO]) should be provided as required. A soft, easily digestible diet should be continued unless the patient has abdominal distention or ileus. Antibiotic therapy is critical to minimize complications (Table 190-7). It has been suggested that traditional therapy with either chloramphenicol or amoxicillin is associated with relapse rates of 5-15% and 4-8%, respectively, whereas use of the quinolones and third-generation cephalosporins is associated with higher cure rates. The antibiotic treatment of typhoid fever in children is also influenced by the prevalence of antimicrobial resistance. Over the past 2 decades, emergence of multidrug-resistant strains of S. Typhi (i.e., isolates fully resistant to amoxicillin, trimethoprim-sulfamethoxazole, and chloramphenicol) has necessitated treatment with fluoroquinolones, which are the antimicrobial drug of choice for treatment of salmonellosis in adults, or cephalosporins. The emergence of resistance to quinolones has placed tremendous pressure on public health systems because alternative therapeutic options are limited.
Bhutta ZA. Current concepts in the diagnosis and treatment of typhoid fever. Br Med J. 2006;333:78-82.
Crump JA, Barrett TJ, Nelson JT, et al. Reevaluating fluoroquinolone breakpoints for Salmonella enterica serotype Typhi and for non-Typhi salmonellae. Clin Infect Dis. 2003;37:75-81.
Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. Bull WHO. 2004;82:346-353.
Crump JA, Mintz ED. Global trends in typhoid and paratyphoid Fever. Clin Infect Dis. 2010;50:241-246.
Dolecek C, Tran TP, Nguyen NR, et al. A multi-center randomised controlled trial of gatifloxacin versus azithromycin for the treatment of uncomplicated typhoid fever in children and adults in Vietnam. PLoS One. 2008;3:e2188.
Effa EE, Bukirwa H: Azithromycin for treating uncomplicated typhoid and paratyphoid fever (enteric fever) (review), Cochrane Data System Rev (4):CD006083, 2008.
Fangtham M, Wilde H. Emergence of Salmonella paratyphi A as a major cause of enteric fever: need for early detection, preventive measures, and effective vaccines. J Travel Med. 2008;15:344-350.
Gasem MH, Keuter M, Dolmans WM, et al. Persistence of salmonellae in blood and bone marrow: randomized controlled trial comparing ciprofloxacin and chloramphenicol treatments against enteric fever. Antimicrob Agents Chemother. 2003;47:1727-1731.
House D, Bishop A, Parry C, et al. Typhoid fever: pathogenesis and disease. Curr Opin Infect Dis. 2001;14:573-578.
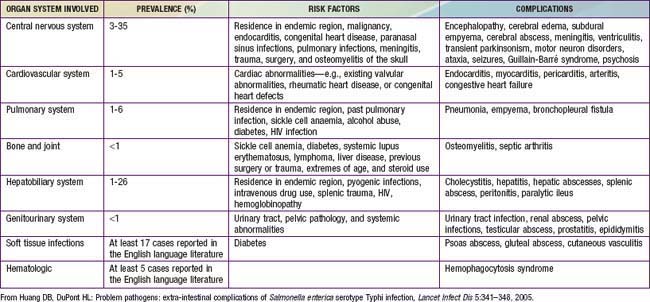
Huang DB, DuPont HL. Problem pathogens: extra-intestinal complications of Salmonella enterica serotype Typhi infection. Lancet Infect Dis. 2005;5:341-348.
Luby SP, Faizan MK, Fisher-Hoch SP, et al. Risk factors for typhoid fever in an endemic setting, Karachi, Pakistan. Epidemiol Infect. 1998;120:129-138.
Lynch MF, Blanton EM, Bulens S, et al. Typhoid fever in the United States, 1999–2006. JAMA. 2009;302:859-865.
Parry CM, Hien TT, Dougan G, et al. Typhoid fever. N Engl J Med. 2002;347:1770-1782.
Prakash P, Mishra OP, Singh AK, et al. Evaluation of nested PCR in diagnosis of typhoid fever. J Clin Microbiol. 2005;43:431-432.
Sinha A, Sazawal S, Kumar R, et al. Typhoid fever in children aged less than 5 years. Lancet. 1999;354:734-737.
Sur D, Ochiai RL, Bhattacharya SK, et al. A cluster-randomized effectiveness trial of Vi typhoid vaccine in India. N Engl J Med. 2009;361:335-344.
Thaver D, Zaidi AK, Critchley J, et al. A comparison of fluoroquinolones versus other antibiotics for treating enteric fever: meta-analysis. BMJ. 2009;338:b1865.
Wain J, House D, Parkhill J, et al. Unlocking the genome of the human typhoid bacillus. Lancet Infect Dis. 2002;2:163-170.
World Health Organization. Background document: The diagnosis, prevention and treatment of typhoid fever. Communicable disease surveillance and response: vaccines and biologicals. World Health Organization, Geneva, 2003;19-23. http://whqlibdoc.who.int/hq/2003/WHO_V&B_03.07.pdf.