CHAPTER 17 Protein Synthesis and Folding*
The nuclear genome contains information specifying many thousands of proteins. Whatever their final destination—nucleus, cytoplasm, membrane-bound organelles, or extracellular space—these proteins are synthesized in the cytoplasm. The few proteins encoded by genes in mitochondria and chloroplasts are synthesized in those organelles. The biochemical synthesis of proteins is called translation, as the process translates sequences of nucleotides in a messenger RNA (mRNA) into the sequence of amino acids in a polypeptide chain. Translation of mRNA requires the concerted actions of small transfer RNAs (tRNAs) linked to amino acids, ribosomes (complexes of RNA and protein), and many soluble proteins. GTP binding and hydrolysis regulate several proteins that orchestrate the interactions of these components. Ultimately, RNA bases in the ribosome catalyze the formation of peptide bonds. Some newly synthesized polypeptides fold spontaneously into their native structure in the cellular environment, but many require assistance from proteins called chaperones. It has been proposed that the bulk of the evolution of the translation apparatus occurred after the basic mechanisms were established, to provide greater precision. This perspective seems to explain the extraordinary complexity of the process.
Protein Synthetic Machinery
Messenger RNA
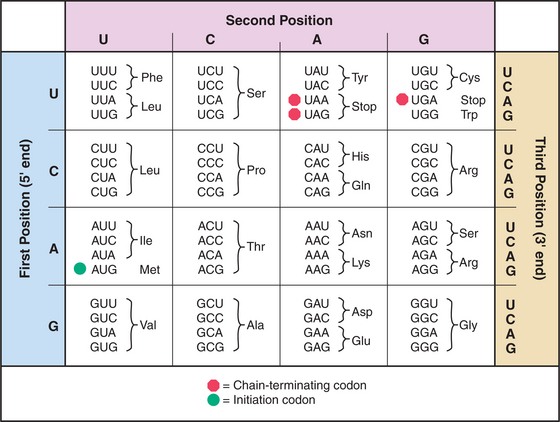
mRNAs have three parts: Nucleotides at the 5′ end provide binding sites for proteins that initiate polypeptide synthesis; nucleotides in the middle specify the sequence of amino acids in the polypeptide; and nucleotides at the 3′ end regulate the stability of the mRNA (see Figs. 15-1 and 16-1). Within the protein-coding region, successive triplets of three nucleotides, called codons, specify the sequence of amino acids. The genetic code relating nucleotide triplets to amino acids is, with a few minor exceptions, universal. One to six different triplet codons encode each amino acid (Fig. 17-1). An initiation codon (AUG) specifies methionine, which begins all polypeptide chains. In addition, any one of three termination codons (UAA, UGA, UAG) stops peptide synthesis.
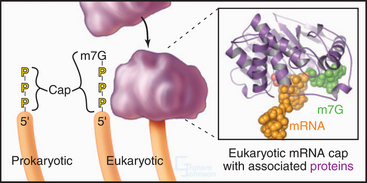
Eukaryotic and bacterial mRNAs differ in three ways. First, eukaryotic mRNAs encode one protein, and bacterial mRNAs generally encode more than one protein. Second, most eukaryotic (and eukaryotic viral) mRNAs are capped by an inverted 7-methylguanosine residue joined onto the 5′ end of the mRNA by a 5′-triphosphate-5′ linkage (Fig. 17-2). This 5′ cap is stable throughout the life of the mRNA and protects the 5′ end against attack by nucleases. Third, most eukaryotic mRNAs have a tail of 50 to 200 adenine residues added posttranscriptionally to the 3′ end (see Fig. 16-3). The poly(A) tail may protect the mRNA from degradation in the cytoplasm and increase reinitiation of transcription. Bacterial mRNAs lack 5′ caps or 3′ poly(A) tails. Most eukaryotic mRNAs require processing to remove introns (see Fig. 16-4). Many single-stranded mRNAs have some secondary structure (see Fig. 3-19) stabilized by hydrogen bonding of complementary bases. This secondary structure must be disrupted during translation to allow reading of each codon.
Transfer RNA
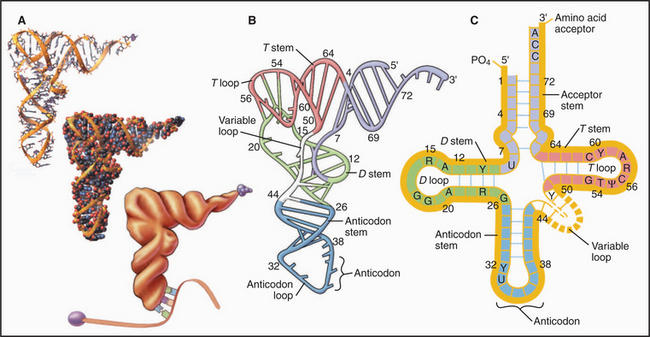
tRNAs are adapters that deliver amino acids to the translation machinery by matching mRNA codons with their corresponding amino acids as they are incorporated into a growing polypeptide (Fig. 17-3). One to four different tRNAs are specific for each amino acid, generally reflecting their abundance in proteins. Specialized tRNAs carrying methionine (formylmethionine in Bacteria) initiate protein synthesis. Transfer RNAs consist of about 76 nucleotides that base-pair to form four stems and three intervening loops. These elements of secondary structure fold to form an L-shaped molecule stabilized by base pairing. A “decoding” triplet (the anticodon) is at one end of the L (the anticodon arm), and the amino acid acceptor site is at the other end of the L (the acceptor arm).
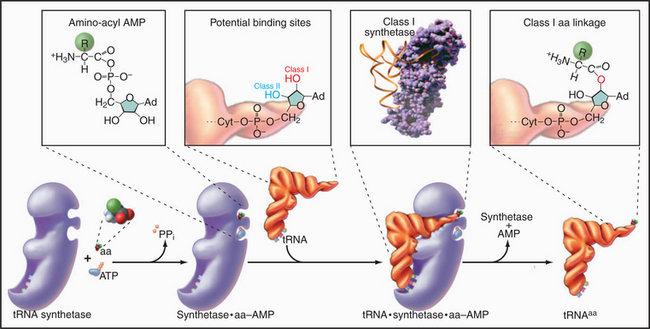
Enzymes called aminoacyl-tRNA (aa-tRNA) synthetases catalyze a two-step reaction that couples an amino acid covalently to its cognate tRNA but not to any other tRNA (Fig. 17-4). In the first step, adenosine triphosphate (ATP) and the amino acid react to form a high-energy aminoacyl adenosine monophosphate (AMP) intermediate with release of pyrophosphate. The second step transfers the amino acid to the 3′ adenine of tRNA, forming an aa-tRNA. This reaction is appropriately called charging, since the high-energy bond between the amino acid and the tRNA activates the amino acid in preparation for forming a peptide bond with an amino group in the growing polypeptide chain. Each of the 20 aa-tRNA synthetases couples a particular amino acid to all of its corresponding tRNAs.
The fidelity of protein synthesis depends on near-perfect coupling of amino acids to the appropriate tRNAs. Synthetases make this selection by interacting with as many as three areas of their cognate tRNAs: anticodon, 3′ acceptor stem, and the surface between these sites (Fig. 17-4). To distinguish between appropriate and inappropriate amino acids, synthetases use proofreading steps, which remove incorrectly paired amino acids from tRNAs.
Ribosomes
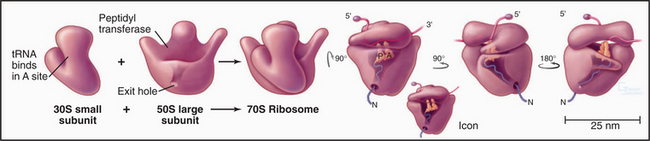
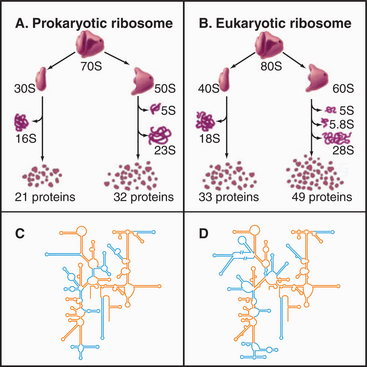
Ribosomes are giant macromolecular machines that bring together an mRNA and aa-tRNAs to synthesize a polypeptide. Base pairing between mRNA codons and tRNA anticodons directs the synthesis of a polypeptide in the order specified by the mRNA codons. Ribosomes consist of a small subunit and a large subunit that bind together during translation of an mRNA (Fig. 17-5). Each subunit consists of one or more ribosomal RNA (rRNA) molecules and many distinct proteins (Fig. 17-6). The sizes of these subunits and rRNAs are traditionally given in units of S, the sedimentation coefficient measured in an ultracentrifuge.
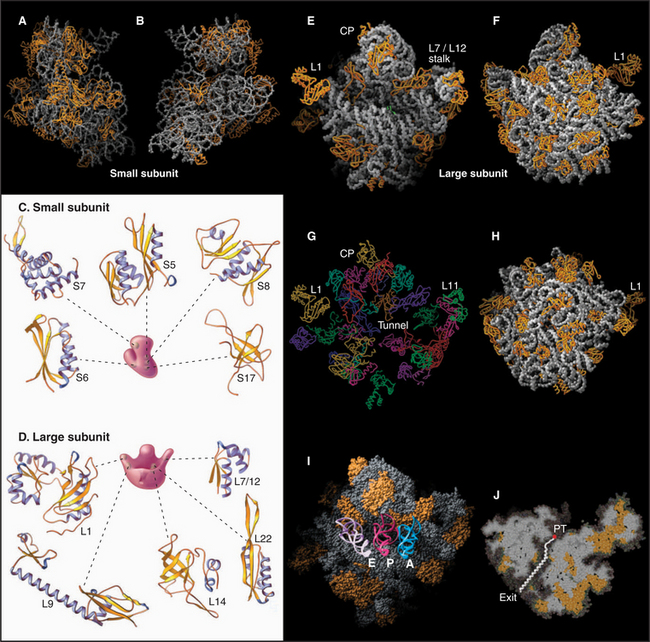
Ribosomal RNAs constitute the structural core of each ribosomal subunit (Fig. 17-7). The 16S rRNA of the small subunit consists of 1500 bases, most of which are folded into base-paired helices. The large subunit contains two RNAs: 23 S rRNA consisting of 2900 bases and 5S rRNA of 121 bases. The rRNAs fold into many based-paired helices, as predicted by phylogenetic analysis of sequences (Fig. 17-6). These helices and their intervening loops pack into a compact structure, as is seen in both surface views and cross sections. Although eukaryotic rRNAs differ in size and sequence from prokaryote rRNAs, their predicted secondary structures are similar, and they are expected to fold in similar ways. Many features of rRNAs have been conserved during evolution, including the surfaces where subunits and elements of RNA structure interact; sites that are required for binding tRNA, mRNA, and protein cofactors; and the residues involved with peptide bond formation.
Most ribosomal proteins associate with the surface of the rRNA core, although several extend peptide strands into the core (Fig. 17-7). Ribosomal proteins are generally small (10 to 30 kD) and basic, but each has a unique structure. With one exception, ribosomes have just one copy of each protein.
Outline of Protein Synthesis
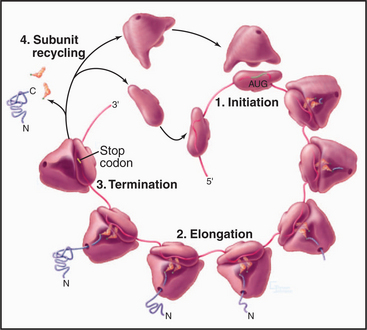
Organisms in all three domains of life use many homologous components and similar reactions for protein synthesis, but many of the details differ as is expected after 3 billion years of evolutionary divergence. In all three domains, protein synthesis takes place in four steps: initiation, elongation, termination, and subunit recycling (Fig. 17-8). Guanosine triphosphatase (GTPase) proteins regulate the progress and fidelity of many of the steps (see Fig. 4-6 for details on GTPase cycles). Initiation, elongation, and termination all depend on directed movement of molecular machinery along an mRNA and precise recognition between amino acids, tRNAs, adapter proteins, and the gene sequence encoded in the mRNA.
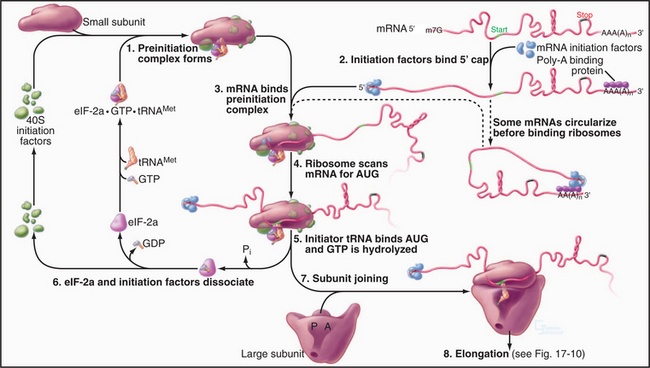
Initiation Phase
The goal of initiation is to bring together the initiator tRNA carrying methionine (or N-formylmethionine, fMet, in Bacteria) and the AUG initiator codon of the mRNA on the ribosome (Fig. 17-9). In eukaryotes, more than 10 soluble protein factors (eukaryotic initiation factors, or eIF) coordinate the interactions of the RNA molecules. Fewer protein factors (designated IF) participate in prokaryotes. In eukaryotes, several steps occur in succession:
Initiation is the most highly regulated step in protein synthesis, frequently involving phosphorylation of initiation factors. For example, phosphorylation increases the affinity of eIF-2a for its guanine nucleotide-exchange factor (eIF-2b). Strongly bound eIF-2b inhibits initiation by competing with initiator tRNA for binding eIF-2a. Cells that are subjected to various stresses utilize phosphorylation of eIF-2a to inhibit translation. In contrast, phosphorylation of eIF-4F favors translation by enhancing the interaction of this initiation factor with the 5′ cap of mRNAs. This modulation of affinity can even influence the selective translation of particular mRNAs, since the 5′ caps of mRNAs vary in affinity for eIF-4F.
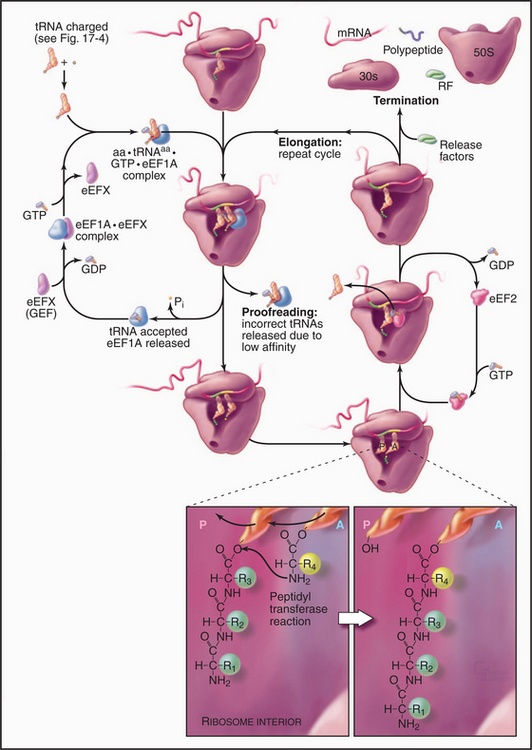
Elongation Phase
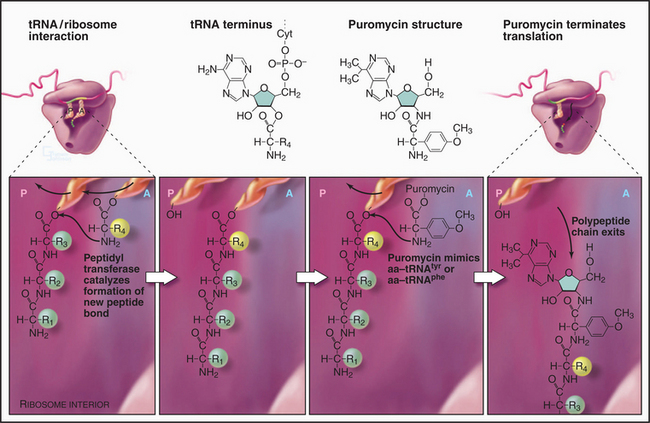
Repetitive cycles of codon-directed incorporation of amino acids into the polypeptide chain begin once the two ribosomal subunits are joined with an initiator tRNA and mRNA properly in place (Fig. 17-10). Each cycle of elongation consists of four steps: (1) binding of an aa-tRNA to the A site on the ribosome; (2) proofreading to ensure that it is the correct aa-tRNA; (3) peptide bond formation; and (4) translocation, which advances the mRNA by one codon and moves the peptidyl-tRNA from the A site to the P site on the ribosome.
The growing peptide threads through a 10-nm-long tunnel in the large subunit lined with RNA (Figs. 17-5, 17-7, and 17-8). The tunnel accommodates an extended polypeptide about 40 residues long. The distal parts of the tunnel are wide enough to pass an a-helix. The N-terminus of longer peptides exits from the large subunit.
Cells balance speed and accuracy during translation to achieve an error rate of about 1 in 104 incorrect amino acids. As a result of this compromise, ribosomes add about 20 amino acids per second to a polypeptide at 37°C, so synthesis of a protein of average size (300 amino acids) takes only 15 seconds. Greater precision might be achieved by slowing translation, but slower cellular growth might be an evolutionary dis-advantage.
Termination Phase
The assembly of a protein stops when a termination codon (UAA, UAG, or UGA) moves into the A site on the small subunit of the ribosome (Fig. 17-10). A release factor called eRF1 (RF1 or RF2 in Bacteria) recognizes stop codons, binds to the A site, and induces the ribosome active site to hydrolyze the peptidyl-tRNA ester in the P site. The completed polypeptide chain threads through the ribosome and is released. The large subunit dissociates from the mRNA and the small subunit, leaving both subunits ready to initiate another round of synthesis. Protein factors might contribute to these recycling reactions, but the details are still being investigated.
Spontaneous Protein Folding
Termination is the final step in translation but just the beginning for a new protein. A polypeptide begins to experience its new environment while still being synthesized. When it is about 40 residues long, its N-terminus emerges from the protected tunnel of the large ribosomal subunit into cytoplasm, where it must fold into a three-dimensional structure (see Fig. 3-5) and find its correct cellular destination.
The structure of folded proteins and the folding mechanism are both encoded in the amino acid sequence, making folding spontaneous under suitable conditions. For the soluble proteins, these conditions are aqueous solvent at physiological temperature, neutral pH, and moderate ionic strength. Folding of transmembrane proteins in a lipid bilayer is quite different (see Chapter 20). In test tube experiments, small soluble proteins can be denatured with high temperature, extremes of pH, or high concentrations of urea or guanidine. Denatured proteins exist as ensembles of unfolded polymers with little residual secondary structure.
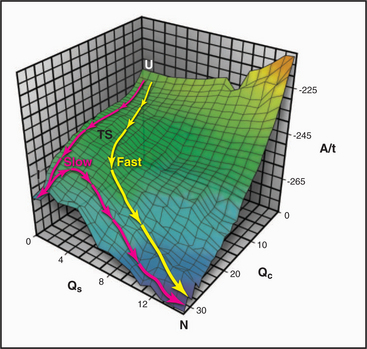
When denatured polypeptides of modest length are transferred to physiological conditions, many fold spontaneously into their native three-dimensional structures on a microsecond to millisecond time scale. (Proteins that require isomerization of prolines, such as collagen, fold much more slowly; see Fig. 29-4.) Starting from many initial denatured states, the polypeptides converge toward a single low-energy native state (Fig. 17-12). The number of possible pathways to the native state is so numerous that if they were sampled individually, proteins would never fold. Thus, both theory and experiment indicate that folding involves a subset of the potential pathways, including an ensemble of loosely folded transition states with elements of secondary structure, certain turns, and hydrophobic contacts found in the core of the native protein.
Misfolding of mutant proteins contributes to many human diseases. For example, the most common cause of cystic fibrosis is genetic deletion of a single amino acid in CFTR, resulting in failure of the protein to fold properly (see Fig. 11-4). Beyond lacking function, misfolded proteins also poison the assembly of native proteins in blistering skin diseases (see Fig. 35-6), hypertrophic cardiomyopathies (see Table 39-4), and other “dominant negative” conditions. Folding of proteins into nonnative states causes prion and amyloid diseases.
Chaperone-Assisted Protein Folding
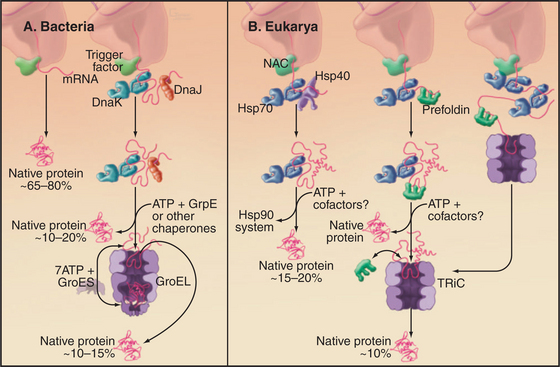
Several families of molecular chaperones (Fig. 17-13) facilitate folding of newly synthesized and denatured proteins. These chaperones do not fold polypeptides by directing the formation of secondary or tertiary structure. Rather, by binding exposed hydrophobic segments of nonnative polypeptides or providing sequestered environments, chaperones inhibit aggregation. They release polypeptides in a folding-competent state for attempts at folding. If folding fails, the cycle of binding and release can be repeated. The following sections cover trigger factor (and other chaperones associated with ribosomes), Hsp70, Hsp90, and cylindrical chaperonins. In addition, specialized chaperones assist with the folding of particular proteins such as tubulin and actin. Mutations in several of these chaperones have been associated with human disease. See Fig. 20-10 for chaperones in the endoplasmic reticulum.
Trigger Factor
Hydrophobic segments of the nascent chain must be protected from aggregation until enough of the chain has emerged from the ribosome to participate in folding. Each growing polypeptide first encounters a chaperone bound next to the exit tunnel on the large ribosomal subunit. The chaperone associated with bacterial ribosomes is called trigger factor (Fig. 17-13). A structurally unrelated protein called nascent polypeptide-associated complex has a similar function in Archaea and eukaryotes. Trigger factor has a hydrophobic groove that is suggested to cradle the growing polypeptide. The signal recognition particle binds on the other side of the exit tunnel, positioned so that its methionine-rich groove (see Fig. 20-5) also interacts with the growing polypeptide. Most bacterial polypeptides fold successfully after being released from trigger factor, while most eukaryotic polypeptides require assistance from additional chaperones.
Hsp70 Chaperones
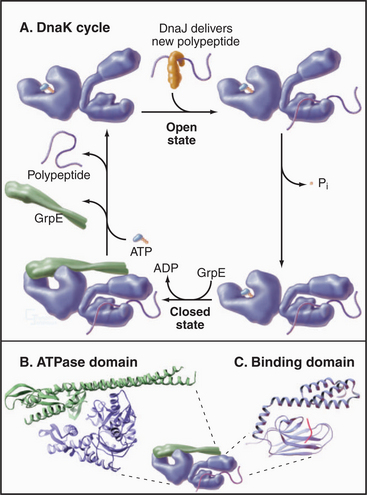
The most widespread chaperones are members of the heat shock protein 70 (Hsp70) family (Fig. 17-14). Their name came from the observation that cells subjected to stresses, such as elevated temperature, increase the synthesis of these proteins to protect against denatured proteins. Hsp70s are present in Archaea, Bacteria (called DnaK), and most compartments of eukaryotes. The family includes Hsp70 in mitochondria and BiP in endoplasmic reticulum (see Fig. 20-4). Budding yeasts have genes for 14 Hsp70s; vertebrates have more.
Hsp70s bind and release peptides with 8 to 13 hydrophobic residues in a wide range of nascent or unfolded polypeptides. ATP binding and hydrolysis drive cycles of peptide binding and release, protecting hydrophobic peptides from aggregation during attempts at folding, delivery to mitochondria and chloroplasts, and import into these organelles (see Figs. 18-4 and 18-6).
Hsp90 Chaperones
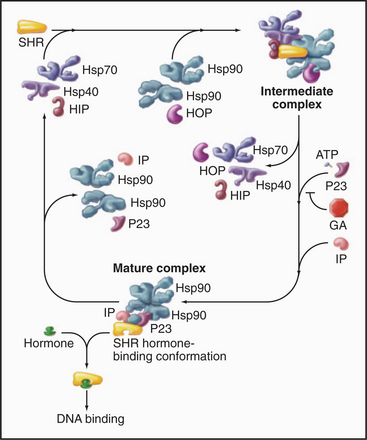
Hsp90 cooperates with other chaperones to stabilize steroid-hormone receptors before they bind their ligands such as progesterone, glucocorticoids, estrogens, or androgens (Fig. 17-15). The chaperones use cycles of ATP hydrolysis to maintain receptors in an “open” state, ready to bind hydrophobic steroids. Steroid binding completes the folding of the receptors and displaces the Hsp90 complex. Then the receptors move to the nucleus to regulate gene expression (see Fig. 15-22). Hsp90 also interacts with other signaling proteins including protein kinases.
Chaperonins
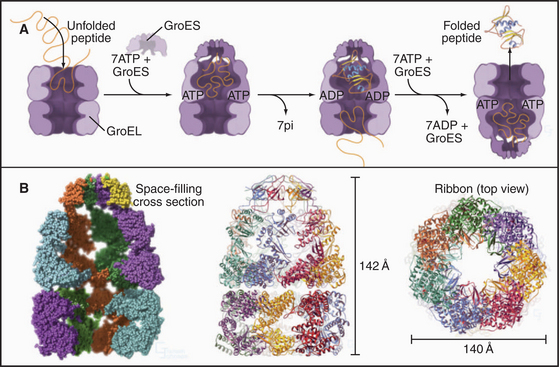
The chaperonin family of barrel-shaped particles promotes efficient protein folding (Fig. 17-16). They allow nascent and denatured polypeptides to fold or refold while sequestered in a cylindrical cavity protected from the complex environment of the cytoplasm. Although 85% of newly synthesized bacterial proteins fold spontaneously or with the assistance of Hsp70s, the remainder require the more isolated folding environment provided by chaperonins (Fig. 17-13). The mechanism of chaperonins is best understood for Escherichia coli GroEL and its co-chaperonin GroES. These assist with folding of nascent polypeptides, which in bacteria occurs largely after translation is complete.
ATP binding and hydrolysis set the tempo for fold-ing cycles. Unfolded polypeptides bind to hydrophobic patches on the inner wall of the GroEL cylinder. Cooperative binding of ATP to each of the subunits in one of the two rings of seven changes their conformation (compare the upper and lower rings in Fig. 17-16B), expanding the internal volume by twofold and favoring binding of a heptameric ring of 10-kD GroES subunits. This closes the top of the cylinder and creates a folding cavity for proteins up to about 70 kD. After ATP hydrolysis on the ring surrounding the folding protein and ATP binding to the opposite ring of seven GroEL subunits, the GroES cap releases, and the cage opens. Folded polypeptides escape into the bulk solution, whereas incompletely folded intermediates can rebind GroEL for another attempt at folding.
Abbott CM, Proud CG. Translation factors: In sickness and in health. Trends Biochem Sci. 2004;29:25-31.
Andersen GR, Nissen P, Nyborg J. Elongation factors in protein biosynthesis. Trends Biochem Sci. 2003;28:434-441.
Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev of Biochem. 2006;75:333-366.
Daggett V, Fersht AR. Is there a unifying mechanism for protein folding? Trends Biochem Sci. 2003;28:18-25.
Dobson CM. Protein folding and misfolding. Nature. 2003;426:884-890.
Frydman J. Folding of newly translated proteins in vivo: The role of molecular chaperones. Annu Rev Biochem. 2004;70:603-647.
Ibba M, Curnow AW, Soll D. Aminoacyl-tRNA synthesis: Divergent routes to a common goal. Trends Biochem Sci. 1997;22:39-42.
Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657-704.
May BC, Govaerts C, Prusiner SB, Cohen FE. Prions: So many fibers, so little infectivity. Trends Biochem Sci. 2004;29:162-165.
Mazumder B, Seshadri V, Fox PL. Translational control by the 3′-UTR: The ends specify the means. Trends Biochem Sci. 2003;28:91-98.
Mitra K, Frank J. Ribosome dynamics: Insights from atomic structure modeling into cryoelectron microscopy maps. Annu Rev Biophys Biomolec Struct. 2006;35:299-317.
Moore PB, Steitz TA. The structural basis of large ribosomal subunit function. Annu Rev Biochem. 2003;72:813-850.
Myers JK, Oas TG. Mechanisms of fast protein folding. Annu Rev Biochem. 2002;71:783-815.
Nakamura Y, Ito K. Making sense of mimic in translation termination. Trends Biochem Sci. 2003;28:99-105.
Ogle JM, Carter AP, Ramakrishnan V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem Sci. 2003;28:259-266.
Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75:271-294.
Piper M, Holt C. RNA translation in axons. Annu Rev Cell Devel Biol. 2004;20:505-523.
Rodnina MV, Wintermeyer W. Peptide bond formation on the ribosome: Structure and mechanism. Curr Opin Struct Biol. 2003;13:334-340.
Saibil HR, Ranson NA. The chaperonin folding machine. Trends Biochem Sci. 2002;27:627-632.
Selkoe DJ. Folding proteins in fatal ways. Nature. 2003;426:891-899.
Sonenberg N, Dever TE. Eukaryotic translation initiation factors and regulators. Curr Opin Struct Biol. 2003;13:56-63.
Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci. 2003;28:182-188.
Young JC, Barral JM, Hartl FU. More than folding: Localized functions of cytosolic chaperones. Trends Biochem Sci. 2003;28:541-547.