116 Principles of Human Genetics
Chromosome Structure and Nomenclature
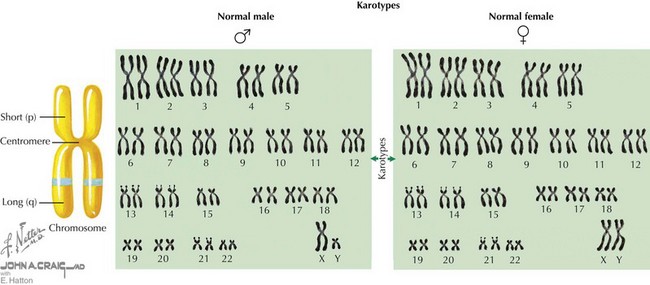
The human genome is composed of deoxyribonucleic acid (DNA), the genetic code for all organisms. Chromosomes are located in the cell nucleus and are composed of DNA packaged with proteins that exist in 23 pairs in every human diploid cell—22 pairs of autosomes (numbered 1-22) and one pair of sex chromosomes (X and Y). Each pair of chromosomes is connected at a region called the centromere, creating two arms for each pair of chromosomes (Figure 116-1). In most chromosomes, the centromere is located closer to one end than the other, forming a short p arm and a longer q arm. Chromosomes can be visualized under a microscope when they are captured during the metaphase stage of the cell cycle. Using a special technique known as Giemsa staining, metaphase chromosomes can be used to determine a karyotype for an individual (see Figure 116-1). A karyotype result indicates the total number of chromosomes identified in the cell, typically 46, followed by the sex chromosomes observed, usually XX or XY. Therefore, a normal female karyotype is 46,XX, and a normal male karyotype is 46,XY.
Methods Of Detecting Aneuploidy
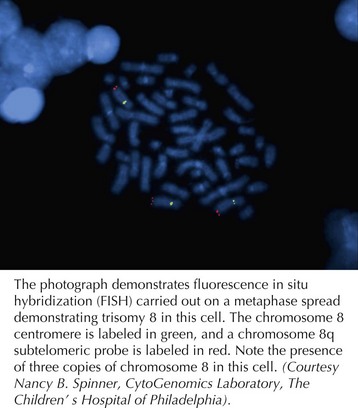
With advances in molecular biology, more specific techniques for evaluating aneuploidy have been developed. In the late 1980s, fluorescent in situ hybridization (FISH) was developed as a method to detect specific chromosomal deletions (Figure 116-2). A fluorescent-labeled DNA sequence that is complementary to a known region of a chromosome is hybridized to a set of chromosomes in a cell. If two copies of the chromosome sequence are present, two fluorescent probes are visualized; if only one probe is seen, it is inferred that one copy of the chromosomal segment is deleted. FISH is used to confirm karyotype findings or to detect deletions too small to be visualized with a standard karyotype. FISH also permits analysis of uncultured cells, permitting diagnoses to be made more rapidly.
Mitosis, Meiosis, and Gamete Formation
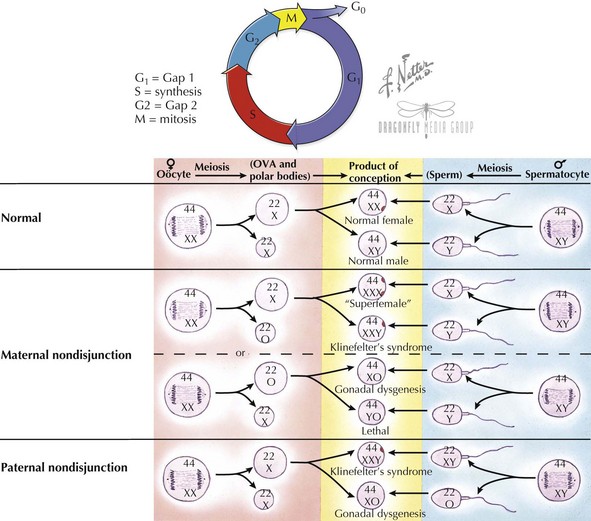
Diploid cells are maintained through the process of mitosis in dividing somatic cell types. Typical cells have four stages to their growth and division cycle (Figure 116-3). The first stage, G1 (for growth 1 or gap 1), is largely composed of growth and has no active cell division. During the next stage (S), DNA is replicated or synthesized to form a four-copied, or tetraploid, genome. After another growth phase (G2) during which the replicated genome is surveyed to correct any aberrations, a division phase (D), during which mitosis occurs, results in two new daughter cells. During mitosis, the replicated tetraploid genome is divided so that each daughter cell receives a diploid complement of chromosomes after cell division. Mitosis is divided into four stages—prophase, metaphase, anaphase, and telophase. At the beginning of prophase, the duplicated chromosomes are elongated and appear as a single chromosome, although two sister chromatids have already been formed by this stage. At the end of prophase, the sister chromatids have become more condensed, and the centrioles are seen at opposite poles of the nucleus. During metaphase, the nuclear membrane disintegrates, and the formation of the mitotic spindle made up of fibers that connect each chromatid of a sister chromatid pair to opposite centrioles, causing them to align in the middle of the nucleus. The centromere of each chromosome also divides in preparation for migration of sister chromatids to opposite centriole spindles. It is during metaphase that chromosomes are typically viewed under the microscope because this is when the chromosomes are most condensed and the mitotic spindle can be arrested with the chemical colchicine. During anaphase, sister chromatics are segregated and migrate toward opposite poles along the spindle. Telophase is marked by the arrival of chromosomes at opposite poles, the disintegration of the spindle, and reformation of a nuclear membrane.
Haploid cells are generated via a special type of germ cell division called meiosis, so that each egg or sperm contributes half of the genetic material to an embryo (see Figure 116-3). A haploid complement of chromosomes consists of one of every autosome (a non-sex chromosome) and an X or Y. In this way, each parent contributes 23 chromosomes to produce a full complement of 46 in an embryo. Meiosis also permits some exchange of genetic material, thus creating genetic variability between gametes. Recombination events, usually at least one per chromosome, result in no two haploid gametes being identical. Recombination events are thought to occur twice as frequently in females as in males. Meiosis is divided into two stages, meiosis I and II. In meiosis I, similar to mitosis, a cell replicates its genome and then divides to create two cells with a full complement of 23 chromosome pairs. In meiosis II, daughter chromatids are divided among daughter cells, such that each contains a single copy of each of the 22 autosomes and an X or Y sex chromosome. In males, all four resulting haploid cells differentiate into sperm. In females, only one of these four haploid set of chromosomes ends up as an ova; the other products become polar bodies. A common cause of aneuploidy is thought to arise from nondisjunction, or failure of the chromosomes to divide equally between daughter cells during meiosis.
Gene Structure
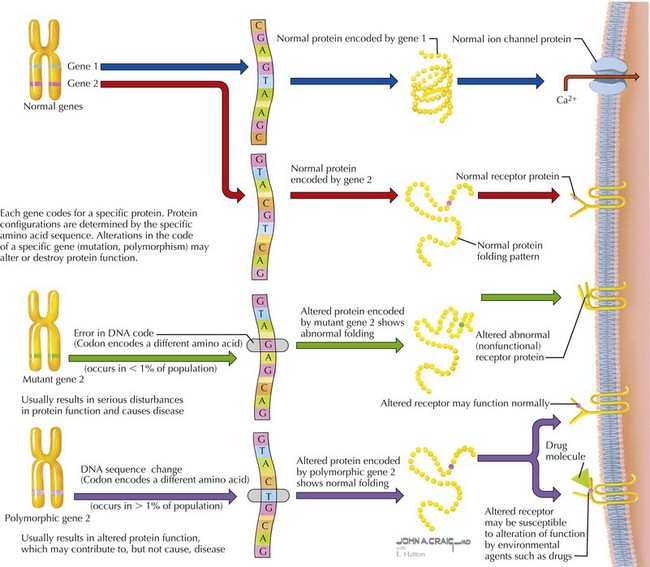
DNA is a double-stranded macromolecule composed of complementary pairs of the purine bases, adenosine and guanine, and the pyrimidine bases, thymine and cytosine (Figure 116-4). The entire haploid genome is composed of some 3 billion base pairs, with each chromosome composed of 50 to 150 million base pairs. Of the entire genome, only about 2% is coding sequence (i.e., encodes genes that are transcribed into proteins). The remainder of the genome is noncoding sequence presumably involved in structural integrity of chromosomes or control of gene expression. A typical gene is transcribed into RNA (RNA), which in turn is translated into a functional protein composed of amino acids. Elements of the genomic DNA surrounding a gene, known as promoters and enhancers, serve to regulate the location, timing. and level of transcription. Before translation, transcribed RNA is processed to form messenger RNA (mRNA) by removing or splicing out intervening sequences known as introns to result in a mature mRNA containing only exons. Mature mRNA moves from the nucleus to the cytoplasm of a cell, where it can be translated into protein in a structure called the ribosome. The ribosome scans the mRNA codons, triplets of nucleotides that encode a specific amino acid, or a start or stop signal. Using the mRNA as a template, ribosomal RNA (rRNA), ribosomal proteins, and transfer RNA (tRNA), the ribosome delivers the appropriate amino acid into the chain of amino acids in the formation of a translated protein. At any stage in the process of transcription or translation, several types of errors can occur, resulting in alterations of protein structure, function, or both (see Figure 116-4).
Modes of Inheritance
Evaluation and Diagnosis of Genetic Syndromes
Categories of Dysmorphic Disorders
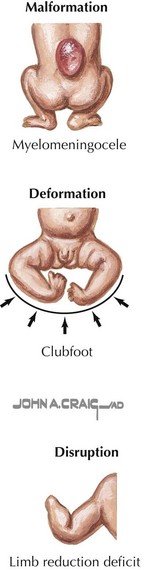
Malformations can also be caused by an isolated disruption or deformation during embryogenesis unrelated to a genetic defect or teratogens (Figure 116-5). Disruption is a mechanical factor that can occur in utero, resulting in an isolated or in a series of structural abnormalities. An example of a disruption event that can lead to congenital anomalies is amniotic band syndrome. Limb and craniofacial defects are thought to arise from either a vascular disruption or strangulation of fetal digits or extremities by strands of a partially disrupted amniotic sac. Deformation describes an abnormal shape or form caused by mechanical forces from within the fetus or outside the fetus in the intrauterine environment. Examples of deformation include clubfoot and congenital dislocation of the hip. Deformations can occur as the result of crowding in the uterus, as in twin gestations.
Evaluation of Children with Dysmorphism
Although genetic testing is often used for definitive diagnosis of a genetic syndrome, other information must be collected to effectively generate a differential diagnosis to guide management and testing. First, a careful history should be obtained that includes the prenatal course, birth history, medical history preceding diagnostic evaluation, and assessment of the motor and cognitive development. Second, a complete family pedigree should be obtained to determine possible heredity or familial risk factors pertaining to a diagnosis. Third, a full physical examination, including anthropomorphic measurements and careful attention to dysmorphic features, is necessary (Table 116-1). With all of this information collected, literature review is often needed to assist in generating an appropriate differential diagnosis. Comparison of findings with similar cases is of particular value with the rarer conditions. Finally, cytogenetic, molecular diagnostic testing, and specific gene testing are used to confirm or exclude a clinical diagnosis.
| System | Features | Examples |
|---|---|---|
| Growth parameters |
Baird PA, Anderson TW, Newcombe HB, et al. Genetic disorders in children and young adults: a population based study. Am J Med Genet. 1988;42:677-693.
Hall JG, Powers EK, Mcllvaine RT, Ean VH. The frequency and financial burden of genetic disease in a pediatric hospital. Am J Med Genet. 1978;1(4):417-436.
Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860-921.
McCandless SE, Brunger JW, Cassidy SB. The burden of genetic disease on inpatient care in a children’s hospital. Am J Hum Genet. 2004;74:121-127.
Scriver CR, Neal JL, Saginur R, et al. The frequency of genetic disease and congenital malformation among patients in a pediatric hospital. Can Med Assoc J. 1973;108:1111-1115.
Stevenson DA, Carey JC. Contribution of malformations and genetic disorders to mortality in a children’s hospital. Am J Med Genet A. 2004;126A(4):393-397.
U.S. Department of Energy. Genomic Science Program. Available at http://genomics.energy.gov
Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470-476.