html xmlns=”http://www.w3.org/1999/xhtml”>
Pharmacogenomics
Introduction
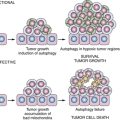
Pharmacogenomics in Oncology
Genotyping and Phenotyping
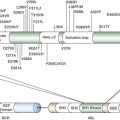
Pharmacogenomic Discovery Approaches
Single-Gene Approaches
Pathway Approaches
Genome-Wide Approaches
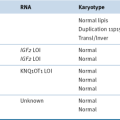
Whole-Genome Sequencing (WGS)
Bioinformatic Considerations
Clinical Relevance of Pharmacogenomic Findings During Implementation
Conclusions
1. Moderne Probleme der Humangenetik . Ergeb Inn Med Kinderheilkd . 1959 ; 12 : 52 – 125 .
2. Pharmacogenetics . Prog Med Genet . 1964 ; 23 : 49 – 74 .
3. Initial sequencing and analysis of the human genome . Nature . 2001 ; 409 : 860 – 921 .
4. The sequence of the human genome . Science . 2001 ; 291 : 1304 – 1351 .
5. Preparing for precision medicine . N Engl J Med . 2012 ; 366 : 489 – 491 .
6. Genomics and drug response . N Engl J Med . 2011 ; 364 : 1144 – 1153 .
7. Pharmacogenomic contribution to drug response . Cancer J . 2011 ; 17 : 80 – 88 .
8. Combining immunotherapy and targeted therapies in cancer treatment . Nat Rev Cancer . 2012 ; 12 : 237 – 251 .
9. Germline pharmacogenomics in oncology: decoding the patient for targeting therapy . Mol Oncol . 2012 ; 6 : 251 – 259 .
10. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy . Nature . 2005 ; 434 : 917 – 921 .
11. The potential of exploiting DNA-repair defects for optimizing lung cancer treatment . Nat Rev Clin Oncol . 2012 ; 9 : 144 – 155 .
12. Cancer genomics: technology, discovery, and translation . J Clin Oncol . 2012 ; 30 : 647 – 660 .
13. Clopidogrel: a case for indication-specific pharmacogenetics . Clin Pharmacol Ther . 2012 ; 91 : 774 – 776 .
14. New natural products in cancer chemotherapy . J Clin Pharmacol . 1990 ; 30 : 770 – 788 .
15. A clinical study of a camptothecin derivative, CPT-11, on hematological malignancies . Proc Am Soc Clin Oncol . 1989 ; 8 : 261 .
16. CPT-11 converting enzyme from rat serum: purification and some properties . J Pharmacobiodyn . 1991 ; 14 : 341 – 349 .
17. Identification of the metabolites of irinotecan, a new derivative of camptothecin, in rat bile and its biliary excretion . Xenobiotica . 1991 ; 21 : 1159 – 1169 .
18. Metabolic fate of irinotecan in humans: correlation of glucuronidation with diarrhea . Cancer Res . 1994 ; 54 : 3723 – 3725 .
19. Pharmacogenetics of acute azathioprine toxicity: relationship to thiopurine methyltransferase genetic polymorphism . Clin Pharmacol Ther . 1989 ; 46 : 149 – 154 .
20. Severe 5-fluorouracil toxicity secondary to dihydropyrimidine dehydrogenase deficiency. A potentially more common pharmacogenetic syndrome . Cancer . 1991 ; 68 : 499 – 501 .
21. Paradoxical relationship between acetylator phenotype and amonafide toxicity . Clin Pharmacol Ther . 1991 ; 50 : 573 – 579 .
22. Genetic predisposition to the metabolism of irinotecan (CPT-11). Role of uridine diphosphate glucuronosyltransferase isoform 1A1 in the glucuronidation of its active metabolite (SN-38) in human liver microsomes . J Clin Invest . 1998 ; 101 : 847 – 854 .
23. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert’s syndrome . N Engl J Med . 1995 ; 333 : 1171 – 1175 .
24. Genetic variation in bilirubin UPD-glucuronosyltransferase gene promoter and Gilbert’s syndrome . Lancet . 1996 ; 347 : 578 – 581 .
25. Racial variability in the UDP-glucuronosyltransferase 1 (UGT1A1) promoter: a balanced polymorphism for regulation of bilirubin metabolism? Proc Natl Acad Sci U S A . 1998 ; 95 : 8170 – 8174 .
26. Pharmacogenetics of irinotecan: clinical perspectives on the utility of genotyping . Pharmacogenomics . 2006 ; 7 : 1211 – 1221 .
27. Phenotype-genotype correlation of in vitro SN-38 (active metabolite of irinotecan) and bilirubin glucuronidation in human liver tissue with UGT1A1 promoter polymorphism . Clin Pharmacol Ther . 1999 ; 65 : 576 – 582 .
28. UGT1A1∗28 polymorphism as a determinant of irinotecan disposition and toxicity . Pharmacogenomics J . 2002 ; 2 : 43 – 47 .
29. Genetic variants in the UDP-glucuronosyltransferase 1A1 gene predict the risk of severe neutropenia of irinotecan . J Clin Oncol . 2004 ; 22 : 1382 – 1388 .
30. Irinotecan pharmacokinetics/pharmacodynamics and UGT1A genetic polymorphisms in Japanese: roles of UGT1A1∗6 and ∗28 . Pharmacogenet Genomics . 2007 ; 17 : 497 – 504 .
31. Food and Drug Administration. Revised Label for Camptosar. In Edition 2006.
32. The role of UGT1A1∗28 polymorphism in the pharmacodynamics and pharmacokinetics of irinotecan in patients with metastatic colorectal cancer . J Clin Oncol . 2006 ; 24 : 3061 – 3068 .
33. UGT1A1∗28 genotype and irinotecan-induced neutropenia: dose matters . J Natl Cancer Inst . 2007 ; 99 : 1290 – 1295 .
34. Pharmacogenetic tests in cancer chemotherapy: what physicians should know for clinical application . J Pathol . 2011 ; 223 : 15 – 27 .
35. Comprehensive analysis of UGT1A polymorphisms predictive for pharmacokinetics and treatment outcome in patients with non-small-cell lung cancer treated with irinotecan and cisplatin . J Clin Oncol . 2006 ; 24 : 2237 – 2244 .
36. Predictive role of the UGT1A1, UGT1A7, and UGT1A9 genetic variants and their haplotypes on the outcome of metastatic colorectal cancer patients treated with fluorouracil, leucovorin, and irinotecan . J Clin Oncol . 2009 ; 27 : 2457 – 2465 .
37. Strategy for detecting susceptibility genes with weak or no marginal effect . Hum Hered . 2007 ; 63 : 85 – 92 .
38. Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukemia . JAMA . 2009 ; 301 : 393 – 403 .
39. Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing . Clin Pharmacol Ther . 2011 ; 89 : 387 – 391 .
40. Thiopurine S-methyltransferase: a genetic polymorphism that affects a small number of drugs in a big way . Pharmacogenetics . 2002 ; 12 : 421 – 423 .
41. Food and Drug Administration. Table of Pharmacogenomic Biomarkers in Drug Labels. In Edition 2012. http://www.fda.gov/%20drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm.
42. Preventing toxicity with a gene test . Science . 2003 ; 302 : 588 – 590 .
43. Expression of functional interleukin-15 receptor and autocrine production of interleukin-15 as mechanisms of tumor propagation in multiple myeloma . Blood . 2000 ; 95 : 610 – 618 .
44. High interleukin-15 expression characterizes childhood acute lymphoblastic leukemia with involvement of the CNS . J Clin Oncol . 2007 ; 25 : 4813 – 4820 .
45. Human genome sequencing in health and disease . Annu Rev Med . 2012 ; 63 : 35 – 61 .
46. Methods for handling multiple testing . Adv Genet . 2008 ; 60 : 293 – 308 .
47. Controlling false discoveries in genetic studies . Am J Med Genet B Neuropsychiatr Genet . 2008 ; 147B : 637 – 644 .
48. Interpreting P values in pharmacogenetic studies: a call for process and perspective . J Clin Oncol . 2007 ; 25 : 4513 – 4515 .
49. Replicating genotype-phenotype associations . Nature . 2007 ; 447 : 655 – 660 .
50. Variation in the gene encoding the serotonin 2A receptor is associated with outcome of antidepressant treatment . Am J Hum Genet . 2006 ; 78 : 804 – 814 .
51. Association of GRIK4 with outcome of antidepressant treatment in the STAR∗D cohort . Am J Psychiatry . 2007 ; 164 : 1181 – 1188 .
52. Personalised medicine: not just in our genes . BMJ . 2012 ; 344 : e2161 .
53. PharmGKB: the Pharmacogenetics Knowledge Base . Nucleic Acids Res . 2002 ; 30 : 163 – 165 .