Chapter 56 Pediatric Pharmacogenetics, Pharmacogenomics, and Pharmacoproteomics
Interindividual variability in the response to similar doses of a given medication is an inherent characteristic of adult and pediatric populations. The role of genetic factors in drug disposition and response, pharmacogenetics, has resulted in many examples of how variations in human genes can lead to interindividual differences in pharmacokinetics and drug response at the level of individual patients. Just as in adults, pharmacogenetic variability contributes to the broad range of drug responses observed in children at any given age or developmental stage. Therefore, it is expected that children will benefit from the promise of personalized medicine: identifying the right drug for the right patient at the right time (see Fig. 56-1 on the Nelson Textbook of Pediatrics website at ![]() www.expertconsult.com). However, pediatricians are keenly aware that children are not merely small adults. Numerous maturational processes occur from birth through adolescence, and using information resulting from the Human Gene Project and related initiatives must take into account the changing patterns of gene expression that occur over development to improve pharmacotherapeutics in children.
www.expertconsult.com). However, pediatricians are keenly aware that children are not merely small adults. Numerous maturational processes occur from birth through adolescence, and using information resulting from the Human Gene Project and related initiatives must take into account the changing patterns of gene expression that occur over development to improve pharmacotherapeutics in children.
Pharmacogenetics, Pharmacogenomics, and the Concept of Personalized Medicine
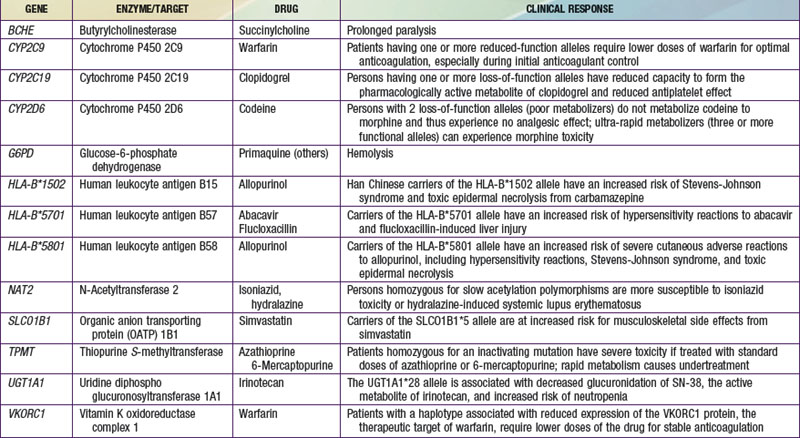
The terms pharmacogenomics and pharmacogenetics tend to be used interchangeably, and precise consensus definitions are often difficult to determine. Pharmacogenetics classically is defined as the study or clinical testing of genetic variations that give rise to interindividual response to drugs. The earliest examples of pharmacogenetic traits include specific adverse drug reactions, such as unusually prolonged respiratory muscle paralysis caused by succinylcholine, hemolysis associated with antimalarial therapy, and isoniazid-induced neurotoxicity, all of which were found to be a consequence of inherited variations in enzyme activity. The importance of pharmacogenetic differences is exemplified by the fact that the half-lives of several drugs are more similar in monozygotic twins than in dizygotic twins. However, in addition to pharmacogenetic differences, environmental factors (diet, smoking status, concomitant drug or toxin exposure), physiologic variables (age, sex, disease, pregnancy), and patients’ compliance all contribute to variations in drug metabolism and response. Ethnicity is another potential genetic determinant of drug variability. Chinese patients who are HLA-B*1502 positive have an increased risk of carbamazepine-induced Stevens-Johnson syndrome; white patients who are HLA-B*5701 positive have an increased risk of hypersensitivity to abacavir (Table 56-1).
Pharmacokinetics describes what the body does to a drug. It is often studied in conjunction with pharmacodynamics, which explores what a drug does to the body. The pharmacokinetic properties of a drug are determined by the genes that control the drug’s disposition in the body (absorption, distribution, metabolism, excretion [ADME]). Drug-metabolizing enzymes and drug transporters play a particularly important role in this process (Table 56-2), and the functional consequences of genetic variations in many drug-metabolizing enzymes have been described among subjects of similar and different ethnic groups. The most common clinical manifestation of pharmacogenetic variability in drug biotransformation is an increased risk of concentration-dependent toxicity resulting from reduced clearance and consequent drug accumulation. An equally important manifestation of this variability is lack of efficacy resulting from variations in metabolism of prodrugs. The pharmacogenetics of drug receptors and other target proteins involved in signal transduction or disease pathogenesis can also be expected to contribute significantly to interindividual variability in drug disposition and response.
Definition of Pharmacogenetics Terms
Genetic polymorphisms (variations) result when copies of a specific gene in a population do not have identical nucleotide sequences. The term allele refers to one of a series of alternative DNA sequences for a particular gene. In humans, there are two copies of every gene. An individual’s genotype for a given gene is determined by the set of alleles that the individual possesses. The most common form of genetic variation involves a single base change at a given location, referred to as a single-nucleotide polymorphism (SNP) (Chapters 72 and 74).
The term genotype refers to an individual’s genetic constitution, whereas the observable characteristics or physical manifestations constitute the phenotype, which is the net consequence of genetic and environmental effects (Chapters 72–77). Pharmacogenetics focuses on the phenotypic consequences of allelic variation in single genes.
Developmental or Pediatric Pharmacogenetics and Pharmacogenomics
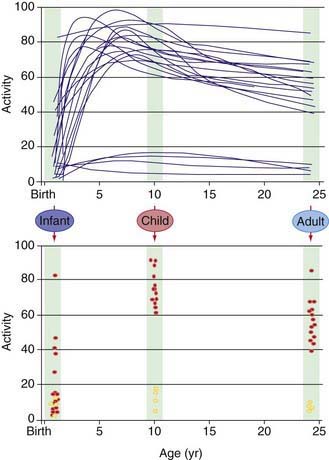
Our understanding of pharmacogenetic principles involves enzymes responsible for drug biotransformation. Individuals are classified as being “fast,” “rapid,” or “extensive” metabolizers at one end of the spectrum, and “slow” or “poor” metabolizers at the other end. This might or might not also include an “intermediate” metabolizer group, depending on the particular enzyme. With regard to biotransformation, children are more complex than adults because fetuses and newborns may be phenotypically slow or poor metabolizers for certain drug-metabolizing pathways because of their stage of development, and they can acquire a phenotype consistent with their genotype at some point later in the developmental process as they mature. Examples of drug-metabolizing pathways that are significantly affected by ontogeny include glucuronidation and some activities of the cytochrome P450 isoenzymes (CYPs) (Chapter 57). It is also apparent that not all infants acquire drug metabolism activity at the same rate. This is due to interactions between genetics and environmental factors. Interindividual variability in the trajectory (i.e., rate and extent) of acquired drug biotransformation capacity may be considered a developmental phenotype (Fig. 56-2), and it helps to explain the considerable variability in some CYP activities observed immediately after birth.
Pharmacogenetic, Pharmacogenomic, Pharmacoproteomic, and Metabolomic Tools
Several genotyping platforms are approved by the U.S. Food and Drug Administration (FDA) and are entering the clinical arena. The Roche Amplichip CYP450 Test was the first such device to receive FDA approval, and at least 6 additional products have since received approval. In general, applications are limited to 1 or 2 genes, such as CYP2C9 and VKORC1 genotyping to guide warfarin therapy or genotyping of UGT1A1 to reduce the risk of irinotecan toxicity. A more comprehensive chip that covers >90% of the ADME markers as defined by the PharmaADME group (http://pharmaadme.org) is available for drug development and research purposes.
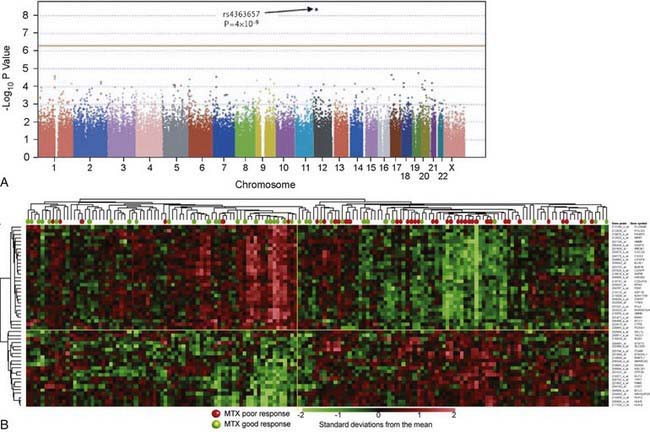
In contrast to pharmacogenetic studies that typically target single genes, pharmacogenomic analyses are considerably broader in scope and focus on complex and highly variable drug-related phenotypes, with targeting of many genes. Genome-wide genotyping technologies make it possible to evaluate genetic variation at more than a million sites throughout an individual genome for SNP and CNV analyses using a single chip. One goal of this type of study is to identify novel genes involved in disease pathogenesis that can lead to new therapeutic targets. Genome-wide association studies (GWAS) are also being applied to identify genetic associations with response to drugs, such as warfarin and clopidogrel, and risk for drug-induced toxicity, including statin-induced myopathy and flucloxacillin hepatotoxicity. The Manhattan plot, a form of data presentation for GWAS, is one way to represent these data (Fig. 56-3A).
Investigating differential gene expression before and after drug exposure has the potential to correlate gene expression with variable drug responses and possibly uncover the mechanisms of tissue-specific drug toxicities. These types of studies use microarray technology to monitor global changes in expression of thousands of genes (the transcriptome) simultaneously. The underlying hypothesis of these global gene-profiling studies is that the measured intensity for each arrayed gene represents its relative expression level. Gene expression profiling data are used to improve disease classification and risk stratification and are used commonly in oncology. For example, this approach has been widely used to address treatment resistance in acute lymphoblastic leukemia and has provided clinically relevant insights into the mechanistic basis of drug resistance and the genomic basis of interindividual variability in drug response. Subsets of transcripts, or gene expression signatures, are being investigated as potential prognostic indicators for identifying patients at risk for treatment failure (Fig. 56-3B).
Developmental Pharmacogenetics of Drug Biotransformation: Applications to Pediatric Drug Therapy Practice
The major consequence of pharmacogenetic polymorphisms in drug-metabolizing enzymes is concentration-dependent toxicity resulting from impaired drug clearance. In certain cases, reduced conversion of prodrug to therapeutically active compounds is also of clinical importance (see Table 56-2). Chemical modification of drugs via biotransformation reactions generally results in termination of biologic activity through decreased affinity for receptors or other cellular targets as well as more rapid elimination from the body. The process of drug biotransformation can be very complex, but it is characterized by 3 important features. First is the concept of broad substrate specificity: A single isozyme can metabolize a large variety of chemically diverse compounds. Second, many different enzymes may be involved in the biotransformation of a single drug (enzyme multiplicity). Finally, a given drug can undergo several different types of reactions. One example of this product multiplicity occurs with racemic warfarin, where at least 7 different hydroxylated metabolites are produced by different CYP isoforms.
Drug biotransformation reactions are conveniently classified into 2 main types, phase I and phase II reactions, which occur sequentially and serve to terminate biologic activity and enhance elimination (Chapter 57). Phase I reactions introduce or reveal (via oxidation, reduction, or hydrolysis) a functional group within the substrate drug molecule that serves as a site for a phase II conjugation reaction. Phase II reactions involve conjugation with endogenous substrates, such as acetate, glucuronic acid, glutathione, glycine, and sulfate. These reactions further increase the polarity of an intermediate metabolite, make the compound more water soluble, and thereby enhance its renal excretion. Interindividual variability in drug biotransformation activity (for both phase I and phase II reactions) is a consequence of the complex interplay among genetic (genotype, sex, race, or ethnic background) and environmental (diet, disease, concurrent medication, other xenobiotic exposure) factors. The pathway and rate of a given compound’s biotransformation is a function of each individual’s unique phenotype with respect to the forms and amounts of drug-metabolizing enzymes expressed.
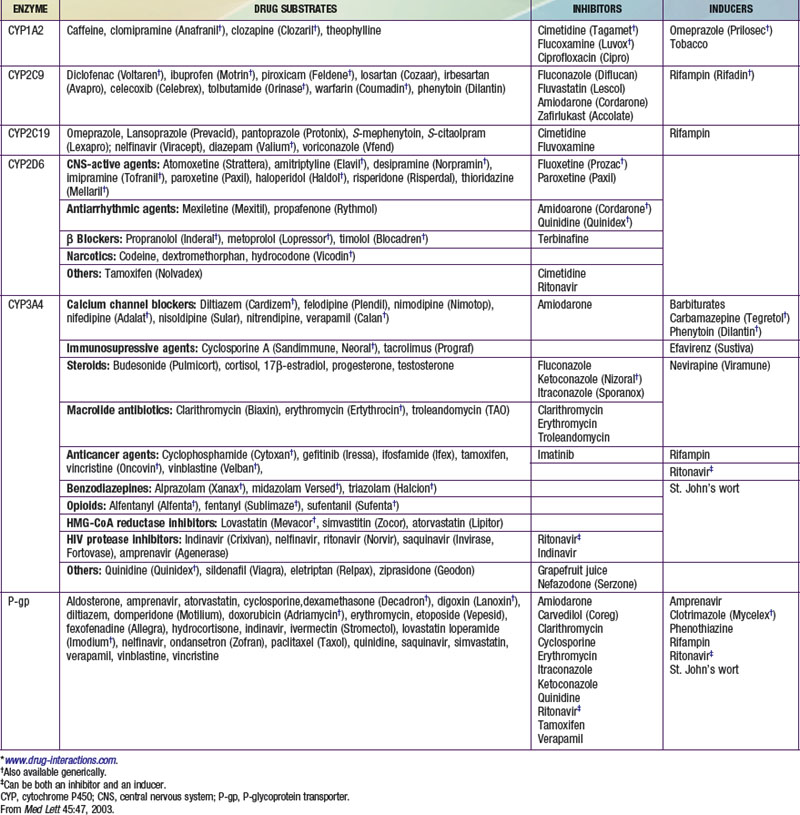
The CYPs are quantitatively the most important of the phase I enzymes. These heme-containing proteins catalyze the metabolism of many lipophilic endogenous substances (steroids, fatty acids, fat-soluble vitamins, prostaglandins, leukotrienes, and thromboxanes) as well as exogenous compounds, including a multitude of drugs and environmental toxins. CYP nomenclature is based on evolutionary considerations. CYPs that share at least 40% homology are grouped into families denoted by an Arabic numeral after the CYP root. Subfamilies, designated by a letter, appear to represent clusters of highly related genes. Members of the human CYP2 family, for example, have >67% amino acid sequence homology. Individual CYPs in a subfamily are numbered sequentially (e.g., CYP3A4, CYP3A5). CYPs that have been identified as being important in human drug metabolism are predominantly found in the CYP1, CYP2, and CYP3 gene families. Importantly, enzyme activity may be induced or inhibited by various agents (see Table 56-2).
CYP2D6
The CYP2D6 gene locus is highly polymorphic, with >75 allelic variants identified to date (http://www.imm.ki.se/CYPalleles/cyp2d6.htm; see Table 56-2). Individual alleles are designated by the gene name (CYP2D6) followed by an asterisk, and an Arabic number. By convention, CYP2D6*1 designates the fully functional wild-type allele. Allelic variants are the consequence of point mutations, single base pair deletions or additions, gene rearrangements, or deletion of the entire gene, resulting in a reduction or complete loss of activity. Inheritance of 2 recessive loss-of-function alleles results in the poor-metabolizer phenotype, which is found in approximately 5-10% of white subjects and approximately 1-2% of Asian subjects. In white subjects, the *3, *4, *5, and *6 alleles are the most common loss-of-function alleles and account for approximately 98% of poor-metabolizer phenotypes. In contrast, CYP2D6 activity on a population basis tends to be lower in Asian and African-American populations owing to a lower frequency of nonfunctional alleles (*3, *4, *5, and *6) and a relatively high frequency of population-selective alleles that are associated with decreased activity relative to the wild-type CYP2D6*1 allele. The CYP2D6*10 allele occurs at a frequency of approximately 50% in Asians, whereas CYP2D6*17 and CYP2D6*29 occur at relatively high frequencies in subjects of black African origin.
CYP2D6 is involved in the biotransformation of >40 therapeutic entities, including several β-receptor antagonists, antiarrhythmics, antidepressants, antipsychotics, and morphine derivatives (for an updated list, see http://medicine.iupui.edu/flockhart; see Table 56-2). CYP2D6 substrates commonly encountered in pediatrics include selective serotonin reuptake inhibitors (SSRIs; fluoxetine, paroxetine, sertraline), risperidone, atomoxetine, promethazine, tramadol, and codeine. Further, over-the-counter cold remedies such as dextromethorphan, diphenhydramine, and chlorphenirame, are also CYP2D6 substrates. An analysis of CYP2D6 ontogeny in vitro that used a relatively large number of samples revealed that CYP2D6 protein and activity remain relatively constant after 1 week of age up to 18 yr. Similarly, results from an in vivo longitudinal phenotyping study involving more than 100 infants over the 1st yr of life demonstrated considerable interindividual variability in CYP2D6 activity but no relationship between CYP2D6 activity and postnatal age between 2 weeks and 12 mo of age. A cross-sectional study involving 586 children reported that the distribution of CYP2D6 phenotypes in children was comparable to that observed in adults by at least 10 yr of age. Thus, available in vitro and in vivo data, albeit based on phenotype data rather than information on drug clearance from pharmacokinetic studies, imply that genetic variation is more important than developmental factors as a determinant of CYP2D6 variability in children.
One consequence of CYP2D6 developmental pharmacogenetics may be the syndrome of irritability, tachypnea, tremors, jitteriness, increased muscle tone, and temperature instability in neonates born to mothers receiving SSRIs during pregnancy. Controversy exists as to whether these symptoms reflect a neonatal withdrawal (hyposerotoninergic) state or represent manifestations of serotonin toxicity analogous to the hyperserotoninergic state associated with the SSRI-induced serotonin syndrome in adults (Chapter 100.1). Delayed expression of CYP2D6 (and CYP3A4) in the first few weeks of life is consistent with a hyperserotoninergic state due to delayed clearance of paroxetine and fluoxetine (CYP2D6) or sertraline (CYP3A4) in neonates exposed to these compounds during pregnancy. Decreases in plasma SSRI concentrations and resolution of symptoms would be expected with increasing postnatal age and maturation of these pathways. Given that treatment of a withdrawal reaction may include administration of an SSRI, there is considerable potential for increased toxicity in affected neonates. Resolution of the question whether symptoms are due to withdrawal versus a hyperserotoninergic state is essential for appropriate management of SSRI-induced neonatal adaptation syndromes. Until further data are available, it would be prudent to consider newborns and infants younger than 28 days of age to be CYP2D6 poor metabolizers.
CYP2C9
Although several clinically useful compounds are substrates for CYP2C9 (http://medicine.iupui.edu/flockhart) (see Table 56-2), the effects of allelic variation are most profound for drugs with a narrow therapeutic index, such as phenytoin, warfarin, and tolbutamide. In vitro studies show a progressive increase in CYP2C9 expression from 1-2% of mature levels in the first trimester to approximately 30% at term. Considerable variability (approximately 35-fold) in expression is apparent over the first 5 mo of life, with approximately half of the samples studied exhibiting values equivalent to those observed in adults. One interpretation of these data is that there is broad interindividual variability in the rate at which CYP2C9 expression is acquired after birth, and in general, the ontogeny of CYP2C9 activity in vivo, as inferred from pharmacokinetic studies of phenytoin in newborns, is consistent with the in vitro results. The apparent half-life of phenytoin is prolonged (∼75 hr) in preterm infants, but it decreases to approximately 20 hr in term newborns. By 2 wk of age, the half-life has further declined to 8 hr. The appearance of concentration-dependent (saturable) metabolism of phenytoin, reflecting the functional acquisition of CYP2C9 activity, does not appear until approximately 10 days of age. The maximal velocity of phenytoin metabolism has been reported to decrease from an average of 14 mg/kg/day in infants to 8 mg/kg/day in adolescents, which might reflect changes in the ratio of liver mass to total body mass observed over this period of development, as has been observed for warfarin.
CYP3A4, CYP3A5, and CYP3A7
The CYP3A subfamily consists of 4 members in humans (CYP3A4, 3A5, 3A7, and 3A43) and is quantitatively the most important group of CYPs in terms of human hepatic drug biotransformation. These isoforms catalyze the oxidation of many different therapeutic entities, several of which are of potential importance to pediatric practice (for an updated list, see http://medicine.iupui.edu/flockhart; see Tables 56-2 and 56-3). CYP3A7 is the predominant CYP isoform in fetal liver and can be detected in embryonic liver as early as 50-60 days’ gestation. CYP3A4, the major CYP3A isoform in adults, is essentially absent in fetal liver but increases gradually throughout childhood. Over the first 6 mo of life, CYP3A7 expression exceeds that of CYP3A4, although its catalytic activity toward most CYP3A substrates is rather limited compared with that of CYP3A4. CYP3A4 is also abundantly expressed in intestine, where it contributes significantly to the first-pass metabolism of orally administered drugs that are substrates (e.g., midazolam). CYP3A5 is polymorphically expressed and is present in approximately 25% of adult liver samples studied in vitro.
Table 56-3 INTERNET RESOURCES FOR PHARMACOGENETICS AND PHARMACOGENOMICS*
* All sites were accessible on November 5, 2009.
Several methods have been proposed to measure CYP3A activity. Using these various phenotyping probes, CYP3A4 activity has been reported to vary widely (up to 50-fold) among individuals, but the population distributions of activity are essentially unimodal, and evidence for polymorphic activity has been elusive. Although 20 allelic variants have been identified (http://www.imm.ki.se/CYPalleles/CYP3A4.htm), most occur relatively infrequently and do not appear to be of clinical importance. Of interest to pediatrics is the CYP3A4*1B allele present in the CYP3A4 promoter region. The clinical significance of this allelic variant appears limited with respect to drug biotransformation activity, despite being associated with 2-fold increased activity over the wild-type CYP3A4*1 allele in in vitro assays. Although there does not appear to be an association between the CYP3A4*1B allele and age of menarche, a significant relationship does exist between the number of CYP3A4*1B alleles and the age at onset of puberty, as defined by Tanner breast score. In one study, 90% of 9 yr old girls with a CYP3A4*1B/*1B genotype had a Tanner breast score of ≥2 compared with 56% of CYP3A4*1A/*1B heterozygotes and 40% of girls homozygous for the CYP3A4*1A allele. Because CYP3A4 plays an important role in testosterone catabolism, it is proposed that the estradiol:testosterone ratio may be shifted toward higher values in the presence of the CYP3A4*1B allele and trigger the hormonal cascade that accompanies puberty. Intestinal CYP3A4 activity is inhibited by grapefruit juice and can result in higher levels of the many drugs metabolized by this enzyme; very large quantities of grapefruit juice can also inhibit the hepatic CYP3A4.
Glucuronosyl Transferases
UGT1A1 is the major UGT gene product responsible for bilirubin glucuronidation, and >100 genetic alterations have been reported (see Table 56-3), most of which are rare and are more properly considered mutations rather than gene polymorphisms (Chapters 96 and 349.1). Inheritance of 2 defective alleles is associated with reduced bilirubin-conjugating activity and gives rise to clinical conditions, such as Crigler-Najjar syndrome and Gilbert syndrome. More frequently occurring polymorphisms involve a dinucleotide (TA) repeat in the atypical TATA box of the UGT1A1 promoter. The wild-type UGT1A1*1 allele has 6 repeats (TA6), and the TA5 (UGT1A1*33), TA7 (UGT1A1*28), and TA8 (UGT1A1*34) variants are all associated with reduced activity. UGT1A1*28, the most common variant, is a contributory factor to prolonged neonatal jaundice. This variant is also associated with impaired glucuronidation and thus toxicity of the active metabolite, SN-38 of the chemotherapeutic agent irinotecan. Allelic variations in UGT1A7 and UGT1A9 have also been associated with irinotecan toxicity in adults with colorectal cancer.
Thiopurine S-Methyltransferase
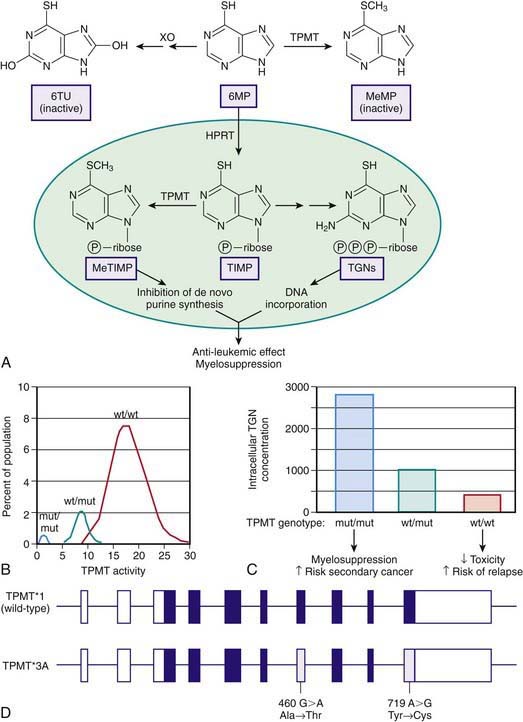
Thiopurine S-methyltransferase (TPMT) is a cytosolic enzyme that catalyses the S-methylation of aromatic and heterocyclic sulfur-containing compounds, such as 6-mercaptopurine (6MP), azathioprine, and 6-thioguanine, used in treating acute lymphoblastic anemia (ALL), inflammatory bowel disease, and juvenile arthritis and for preventing renal allograft rejection. To exert its cytotoxic effects, 6MP requires metabolism to thioguanine nucleotides by a multistep process that is initiated by hypoxanthine guanine phosphoribosyl transferase. TPMT prevents thioguanine nucleotide production by methylating 6MP (Fig. 56-4A). TPMT activity is usually measured in erythrocytes, with activity in erythrocytes reflecting that found in other tissues, including liver and leukemic blasts.
Although approximately 89% of whites and African-Americans have high TPMT activity and 11% have intermediate activity, 1 in 300 persons inherit TPMT deficiency as an autosomal recessive trait (Fig. 56-4B). In newborn infants, peripheral blood TPMT activity is reported to be 50% greater than in race-matched adults and shows a distribution of activity that is consistent with the polymorphism characterized in adults. No data currently indicate how long this higher activity is maintained, although TPMT activities were comparable to previously reported adult values in a population of Korean schoolchildren aged 7-9 yr. In patients with intermediate or low activity, more drug is shunted toward production of cytotoxic thioguanine nucleotides.
TPMT can also methylate 6-thioinosine 5′-monophosphate to generate a methylated metabolite that is capable of inhibiting de novo purine synthesis (Fig. 56-4C). Three mutations have been identified in the TPMT gene (*2, *3A, *3C), which account for 98% of white subjects with low activity. These mutations encode proteins that undergo rapid proteolysis resulting in low enzyme activity.
TPMT*3A is the most common mutant allele and is characterized by 2 nucleotide transition mutations, G460A and A719G, that lead to 2 amino acid substitutions Ala154Thr and Tyr240Cys (Fig. 56-4D). Although the *3A allele only has a frequency of 0.03% in the general population, it represents 55% of all mutant alleles. Either mutation alone results in loss of functional activity through the production of unstable proteins that are subject to accelerated proteolytic degradation. Less-common allelic variants involve SNPs that produce amino acid substitutions in the coding region and defective intron-exon splicing. A polymorphic locus has been identified in the promoter region of the TPMT gene involving 4-8 repeats of a specific nucleotide sequence in tandem. Although these repeats appear to modulate TPMT activity when expressed in vitro, their role in regulating activity in vivo has not been clearly established.
Pharmacogenetics of Drug Transporters
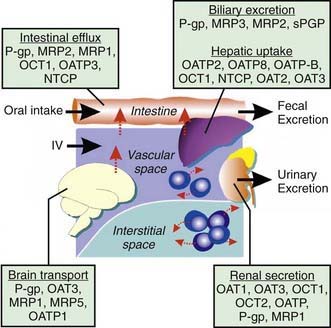
There are several major types of membrane transporters, including organic anion transporter (OATs), organic anion transporting polypeptides (OATPs), organic cation transporting proteins (OCTs), and the adenosine triphosphate (ATP)-binding cassette (ABC) transporters, such as p-glycoprotein and the multidrug-resistant proteins (MRPs). Membrane transporters are heavily involved in drug disposition and actively transport substrate drugs between organs and tissues. Drug transporters are expressed at numerous epithelial barriers, such as intestinal epithelial cells, hepatocytes, renal tubular cells, and at the blood-brain barrier (Fig. 56-5). Transporters often are also determinants of drug resistance, and many drugs work by affecting the function of transporters. Polymorphisms in the genes encoding these proteins can have a significant effect on the ADME and the pharmacodynamic effect of a wide variety of compounds.
Pharmacogenetics of Drug Response: Polymorphisms in Drug Receptors, Ion Channels, and Other Drug Targets during Growth and Development
Drug responses are seldom monogenic events because multiple genes are involved in drug binding to the pharmacologic target and in the subsequent downstream signal transduction events that ultimately collectively manifest as a therapeutic effect. Although genotypes at a particular locus can show a statistically significant effect on the outcome of interest, they might account for only a relatively small amount of the overall population variability for that outcome. For example, a particular group of SNPs in the corticotropin-releasing hormone receptor 1 (CRHR1) gene is associated with a statistically significant improvement in forced expiratory volume in 1 second (FEV1), but accounts for only 6% of the overall variability in response to inhaled corticosteroids (Chapter 138). A series of subsequent studies has determined that allelic variation in several genes in the steroid pathway contributes to overall response to this form of therapy.
The listing and classification of receptors is a major initiative of the International Union of Pharmacology (IUPHAR). The list of receptors and voltage-gated ion channels is available on the IUPHAR website (http://www.iuphar-db.org). The effect of growth and development on the activities and binding affinities of these receptors, effectors, and ion channels has been studied in animals to some extent, but it remains to be elucidated in humans.
Applications for Pharmacogenetics and Pharmacogenomics in Pediatrics
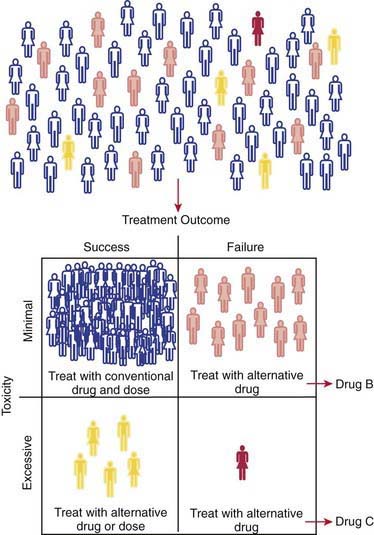
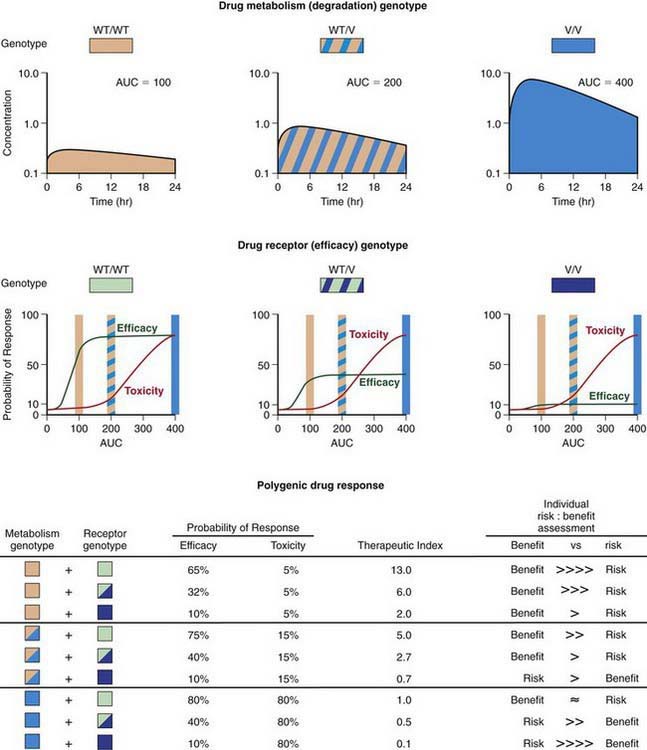
Progress being made in the treatment of ALL provides an outstanding example of how the application of pharmacogenomic principles can improve pediatric drug therapy (Chapter 489). Despite improved understanding of the genetic determinants of drug response, however, many complexities remain to be resolved. Patients with ALL who have 1 wild-type allele and intermediate TPMT activity tend to have a better response to 6MP therapy than patients with 2 wild-type alleles and full activity. Reduced TPMT activity also places patients at risk for irradiation-induced secondary brain tumors and etoposide-induced acute myeloid leukemias. Pharmacogenetic polymorphisms of several additional genes also have the potential to influence successful treatment of ALL. Multiple genetic and treatment-related factors interact to create patient subgroups with varying degrees of risk, and these represent an opportunity for pharmacogenomic approaches to identify subgroups of patients who will benefit from specific treatment regimens and those who will be at risk for short- and long-term toxicities (Fig. 56-6).
2005 AmpliChip CYP450 test. Med Lett Drugs Ther. 2005;47:71-72.
Blake MJ, Gaedigk A, Pearce RE, et al. Ontogeny of dextromethorphan O- and N-demethylation in the first year of life. Clin Pharmacol Ther. 2007;81:510-516.
Chen N, Aleksa K, Woodland C, et al. Ontogeny of drug elimination by the human kidney. Pediatr Nephrol. 2006;21:160-168.
CYP3 A. and drug interactions. Med Lett Drugs. 2005;47:54-55.
Daly AK, Donaldson PT, Bhatnagar P, et al. HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat Genet. 2009;41:816-819.
2004 Drug interactions with grapefruit juice. Med Lett. 2004;46:2-4.
Eckman MH, Rosand J, Greenberg SM, et al. Cost-effectiveness of using pharmacogenetic information in warfarin dosing for patients with nonvalvular atrial fibrillation. Ann Intern Med. 2009;150:73-83.
Eliasson E. Ethnicity and adverse drug reactions. BMJ. 2006;332:1163-1164.
2008 Genetic test for carbamazepine-induced Stevens-Johnson syndrome. Med Lett. 32, 2008.
Hines RN. The ontogeny of drug metabolism enzymes and implications for adverse drug events. Pharmacol Ther. 2008;118:250-267.
Hines RN, McCarver DG. Pharmacogenomics and the future of drug therapy. Pediatr Clin North Am. 2006;53(4):591-619.
Ingelman-Sundberg M. Pharmacogenomic biomarkers for prediction of severe adverse drug reactions. N Engl J Med. 2008;358:637-639.
Koren G, Cairns J, Chitayat D, et al. Pharmacogenetics of morphine poisoning in a breastfed neonate of a codeine-prescribed mother. Lancet. 2006;368:704-705.
Leschziner GD, Andrew T, Pirmohamed M, et al. ABC1 genotype and PGP expression, function and therapeutic drug response: a critical review and recommendations for future research. Pharmacogenomics J. 2007;7:154-179.
O’Donoghue ML, Braunwald E, Antman EM, et al. Pharmacodynamic effect and clinical efficacy of clopidogrel and prasugrel with or without a proton-pump inhibitor: an analysis of two randomized trials. Lancet. 2009;374:989-996.
Perlis RH. Cytochrome P450 genotyping and antidepressants. BMJ. 2007;334:759.
2008 Pharmacogenetic-based dosing of warfarin. Med Lett. 2008;50:39-40.
Schwartz UI, Ritchie MD, Bradford Y, et al. Genetic determinants of response to warfarin during initial anticoagulation. N Engl J Med. 2008;358:999-1008.
Search Collaborative Group. SLCO1B1 variants and statin-induced myopathy—a genomewide study. N Engl J Med. 2008;359:789-799.
Shurin SB, Nabel EG. Pharmacogenomics—ready for prime time? N Engl J Med. 2008;358:1061-1063.
Sorich MJ, Pottier N, Pei D, et al. In vivo response to methotrexate forecasts outcome of acute lymphoblastic leukemia and has a distinct gene expression profile. PLoS Med. 2008;5(4):e83. 15
Stevens JC, Marsh SA, Zaya MJ, et al. Developmental changes in human liver CYP 2D6 expression. Drug Metab Disp. 2008;36:1587-1593.
Takeuchi F, McGinnis R, Bourgeois S, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5(3):e1000433.
Wang L, Weinshilboum R. Thiopurine S-methyltransferase pharmacogenetics: insights, challenges and future directions. Oncogene. 2006;25:1629-1638.
Weiss ST, Litonjua AA, Lange C, et al. Overview of the pharmacogenetics of asthma treatment. Pharmacogenomics J. 2006;6:311-326.
Woodcock J, Lesko L. Pharmacogenetics—tailoring treatment for the outliers. N Engl J Med. 2009;360:811-813.
Yang JJ, Cheng C, Yang W, et al. Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukemia. JAMA. 2009;301:393-403.
Zanger UM, Turpeinen M, Klein K, et al. Functional pharmacogenetics/genomics of human cytochromes P450 involved in drug biotransformation. Anal Bioanal Chem. 2008;392:1093-1108.