CHAPTER 14 Nuclear Structure and Dynamics
The nucleus houses the chromosomes together with the machinery for DNA replication and RNA transcription and processing (Fig. 14-1). Immature RNAs must be kept apart from the translational apparatus because eukaryotic genes are transcribed into RNAs containing noncoding intervening sequences that must be removed by splicing to assemble mature RNA molecules with a continuous open reading frame. Sequestration of immature RNAs is one function of the nuclear envelope, two concentric membrane bilayers that separate the nucleus and cytoplasm. The nuclear envelope also regulates the movement of proteins, such as transcription factors, into and out of the nucleus.
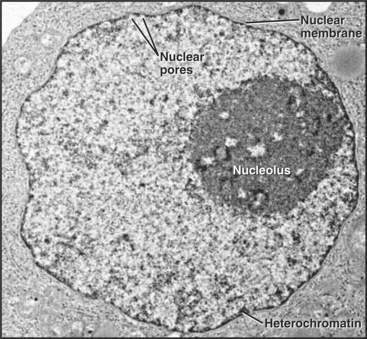
Figure 14-1 ELECTRON MICROGRAPH OF A THIN SECTION OF A NUCLEUS FROM A CANCER CELL WITH THE MAJOR FEATURES LABELED.
(Courtesy of Scott Kaufmann, Mayo Clinic, Rochester, Minnesota.)
This chapter describes what is known about the structure of the nucleus, the nuclear envelope, and the transport of macromolecules into and out of the nucleus. Much is known about the mechanisms of DNA replication (see Chapter 42), RNA transcription and processing (see Chapters 15 and 16), and nuclear trafficking of macromolecules. Currently, less is known about the structural organization of the nucleus and the role of its specialized subcompartments in these nuclear functions.
Overall Organization of the Nucleus
Studies in which entire individual chromosomes are labeled by in situ hybridization (chromosome painting; see Fig. 13-11) reveal that chromosomes tend to occupy discrete regions within the nucleus called chromosome territories. The boundaries of these territories are not absolute, as in some cases an active gene is located well outside its respective territory (see Chapter 13). The poorly defined region between adjacent territories is referred to as the interchromosomal domain. Most RNA transcription, processing, and transport are thought to occur either within or at the boundary of this domain. Although the nucleoplasm is very crowded with chromosomes and RNPs, many nuclear proteins can diffuse rapidly through the nucleus, possibly by moving in the interchromosomal domain.
Specialized Subdomains of the Nucleus
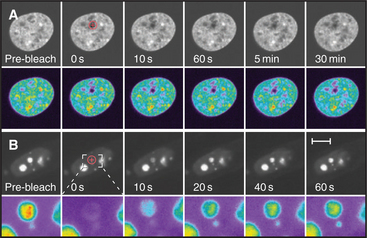
Cell nuclei contain numerous discrete subdomains or bodies with distinctive structural organizations and/or biochemical composition (Fig. 14-2 and Table 14-1). The most prominent of these is the nucleolus, discussed in the next section. Although these subdomains are often referred to as organelles, unlike cytoplasmic organelles, nuclear subdomains are not membrane bounded. In fact, many proteins that have been examined by the fluorescence recovery after photobleaching technique (see Fig. 6-3) exhibit a relatively rapid exchange between a respective body and a nucleoplasmic pool of the protein. Therefore, although these bodies are discrete at steady state, they represent highly dynamic associations of macromolecular complexes. These domains, like the nucleolus, reflect the functional compartmentalization of the nucleoplasm.
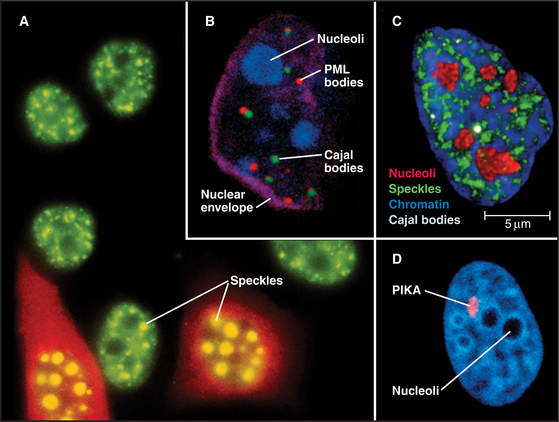
Figure 14-2 examples of major subnuclear structures. A, Components involved in RNA processing are scattered throughout the nucleus but concentrated in domains called speckles that are rich in interchromatin granules. Inhibition of RNA processing causes splicing components to accumulate in enormous concentrations of interchromatin granule clusters. Several cells were injected with a short oligonucleotide that disrupts the function of the U1 snRNP in RNA processing (see Chapter 16), and were then stained with an antibody recognizing the Sm splicing components (green). The injected cells were marked by introducing an inert fluorescent dextran marker into the cytoplasm (red). B, Nucleus with simultaneous staining of nucleoli (blue), PML nuclear bodies (red), Cajal bodies (green), and the nuclear envelope (purple). C, Nucleus with simultaneous staining of chromatin (blue), nucleoli (red), speckles (green), and Cajal bodies (white). D, Nucleus with simultaneous staining of DNA (blue) and the polymorphic interphase karyosomal association (PIKA)/OPT domain (red). Nucleoli appear as unstained areas. A number of proteins involved in the sensing and repair of DNA damage concentrate in the PIKA.
(A, Courtesy of David Spector, Cold Spring Harbor Lab, New York. B–C, Courtesy of Angus Lamond, University of Dundee, Scotland. D, Courtesy of William S. Saunders and William C. Earnshaw.)
| Structure | Comments |
|---|---|
| Cajal bodies | Formerly known as coiled bodies. About 0.2 to 1 μm in diameter, Cajal bodies have a coiled fibrous substructure. First identified by electron microscopy, up to 10 of these structures are seen per cell. They contain a characteristic protein called p80-coilin. They may be involved in snRNP and snoRNP assembly. |
| GEMs | GEMs are usually found paired with Cajal bodies, which they may overlap. They contain the survival of motor neurons (SMN) protein, which is encoded by the gene mutated in spinal muscular atrophy, a severe, inherited, human, muscular wasting disease. SMN and its cofactors appear to play an essential role in the assembly and maturation of snRNPs (see Chapter 16). |
| Nuclear bodies | Function unknown. 5 to 20 spots within the nucleus. Originally observed in electron micrographs of cells following hormonal treatments. However it is not clear that all nuclear bodies described in various cell types are structurally or functionally homologous. A marker antigen for some types of nuclear bodies (called PBC 95K—Mr 95 kD) is recognized by autoantibodies from patients with primary biliary cirrhosis. Some may correspond to PML bodies. |
| Nucleolus | The nucleolus (typically 1 to 5 structures of 0.5 to 5 μm diameter in mammalian cell nuclei) is the site of rRNA transcription and processing, as well as of preribosomal assembly. It is also the site of processing of several other noncoding RNAs, including the RNA component of the signal recognition particle (SRP—Chapter 20). |
| PIKA | The polymorphic interphase karyosomal association (PIKA) was later rediscovered and termed the OPT domain. The PIKA may be up to 5 μm in diameter during G1 phase, but its morphology and number vary across the cell cycle. It appears to correspond to sites of sensing or repair of DNA damage as well as concentrations of certain transcription factors. |
| PML bodies | Also known as PODs and ND10, 10 to 30 of these structures are scattered throughout the nucleus. They are have links with human disease, and in some cases appear to be targeted during viral infections. Fusion of the marker protein PML to the α-retinoic acid receptor is often found in acute promyelocytic leukemia (hence the name PML), in which the PML bodies appear highly fragmented. The link with cancer appears significant, as treatments that are effective against PML appear to restore the normal morphology of PML bodies (see text). |
| Speckles | Speckles are concentrations of components involved in RNA processing. They often correspond to clusters of interchromatin granules seen by electron microscopy. They may serve as storage depots of splicing factors, or they may play a more active role in splicing factor modification and/or assembly. |
When factors involved in RNA processing are detected by fluorescence microscopy, 20 to 50 bright speckles are seen against a diffuse background of nucleoplasmic staining (Fig. 14-2). The diffuse staining probably corresponds to splicing factors associated with perichromatin fibrils at the thousands of sites where RNA transcription and processing take place. Speckles are less prominent in cells that transcribe RNA at high levels, and they become strikingly prominent when RNA processing is inhibited (Fig. 14-2).
When cells enter mitosis, speckles disperse as RNA-processing factors redistribute diffusely throughout the cytoplasm. During telophase, processing factors reaggregate in the cytoplasm into punctate structures termed mitotic interchromatin granule clusters. These factors are subsequently imported into the nucleus at the completion of mitosis.
Cajal bodies (formerly known as coiled bodies) are compact structures about 0.3 to 1.0μm in diameter (Fig. 14-2) that resemble balls of tangled threads in the electron microscope. Nuclei contain 1 to 10 Cajal bodies, which accumulate many factors involved in mRNA processing, as well as a number of nucleolar components, but lack non-snRNP protein-splicing factors that are present in speckles. They also contain an 80-kD human autoantigen of unknown function, called p80-coilin. In contrast to speckles, Cajal bodies disperse when transcription and splicing are blocked, and they are particularly prominent in rapidly growing cells with high levels of gene expression. However, like speckles, some Cajal bodies disassemble during mitosis and reform during the G1 phase after transcription is reinitiated. Cajal bodies are connected with the maturation of newly imported snRNP and snoRNP particles (see Chapter 16). They may also have other functions, including possibly regulating expression of snRNA gene clusters.
Mammalian nuclei also contain about 10 to 30 bodies, varying in size from 0.3 to 1μm, known as promyelocytic leukemia (PML) bodies (other names are listed in Table 14-1) that are often juxtaposed with Cajal bodies (Fig. 14-2). PML bodies were initially defined by the presence of a protein called PML, and other components have since been identified. PML has a RING finger amino acid sequence motif and is therefore likely to be a ligase for ubiquitin or ubiquitin-like proteins (see Fig. 23-8). Its targets are unknown, but several components of PML bodies are conjugated to the ubiquitin-like protein SUMO-1.
The Nucleolus: The Most Prominent Nuclear Subdomain
The nucleolus, first described only five years after the nucleus, in 1835, is the most conspicuous and best-characterized nuclear subdomain (Fig. 14-3). Most mammalian cell nuclei have one to five nucleoli, which are specialized regions 0.5 to 5.0μm in diameter surrounding transcriptionally active ribosomal RNA (rRNA) gene clusters. Within nucleoli occur the bulk of the steps for ribosome biogenesis, from the transcription and processing of rRNA to the initial assembly of ribosomal subunits. The ribosome is a complex macromolecular machine with four different structural RNA molecules and about 85 proteins that are assembled into two subunits (see Figs. 17-6 and 17-7). rRNA transcription by RNA polymerase I comprises nearly one half of total cellular RNA synthesis in some cell types. This high level of synthesis is necessary to produce about 5 million ribosomes in each cell cycle, more than 30 every second in budding yeast.
Over 690 proteins associate stably with human nucleoli. Many more may associate transiently, and this composition changes to reflect different metabolic states of the cell (Fig. 14-4). Many of these nucleolar proteins are involved with either ribosomal RNA synthesis and modification or with ribosome subunit assembly. Surprisingly, the functions of many (˜80) other nucleolar proteins remain unknown and may reflect the involvement of nucleoli in other biological processes. Other stable RNAs, including the RNA component of the signal recognition particle (SRP; see Fig. 20-5), are also processed in the nucleolus, and the nucleolus might have other as yet undiscovered functions.
Ribosomal Biogenesis in Functionally Distinct Regions of the Nucleolus
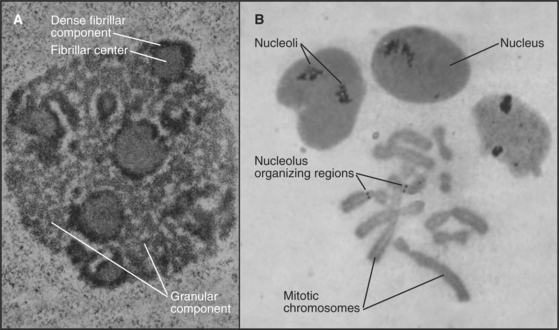
The nucleolus contains three morphologically distinct regions in thin sections viewed by transmission electron microscopy (Fig. 14-3). Fibrillar centers contain concentrations of rRNA genes, together with significant amounts of RNA polymerase I and its associated transcription factors. Actively transcribed ribosomal genes are found near the border between the fibrillar centers and a dense fibrillar component that surrounds them. The granular component is the site for many steps in ribosome subunit assembly and is made up of densely packed clusters of preribosomal particles 15 to 20 nm in diameter.
Ribosomal RNA loci have a modular organization, with genes alternating with spacer regions in large tandemly arranged clusters (see Fig. 16-9). The repeat unit in this array (gene plus spacer) is approximately 40,000 base pairs in humans. Humans have approximately 300 to 400 copies of the ribosomal DNA (rDNA) repeat unit located in clusters on chromosomes 13, 14, 15, 21, and 22. Usually, only a fraction of these genes is actively transcribed. An additional rRNA, 5S, is encoded by distinct genes and transcribed by RNA polymerase III (see Fig. 15-10).
A simple yet efficient mechanism guarantees a balance between the RNA components of the two ribosomal subunits. The major rRNA components are encoded by a single precursor RNA molecule. In humans, this 13,000-base precursor is commonly described by its sedimentation coefficient in sucrose gradients as 45S. Following its transcription, the RNA precursor is processed in a series of cleavages to yield the 18S, 5.8S, and 28S rRNA molecules (see Fig. 16-9). In addition to the cleavages, rRNA processing also involves extensive base and sugar modifications, including approximately 100 2′-O-methyl ribose and approximately 90 pseudouridine residues per molecule. The earliest stages of rRNA processing probably occur in the dense fibrillar component of the nucleolus. Later stages take place in the granular component. Ribosomal protein synthesis occurs in the cytoplasm on free ribosomes, and the newly synthesized proteins are transported into the nucleus for assembly into ribosomes, predominantly in the granular component.
Disassembly of the Nucleolus during Mitosis
The nucleolus disassembles during each mitotic cycle, starting with the dispersal of the dense fibrillar and granular components during prophase. This disassembly is apparently driven by specific phosphorylation of nucleolar proteins. Ultimately, the fibrillar centers alone remain associated with the mitotic chromosomes, forming what are termed nucleolus-organizing regions (NORs [Fig. 14-3B]). NORs are often the sites of a prominent secondary constriction of the chromosome. (The primary constriction is the centromere.) The nucleolar proteins nucleolin and RNA polymerase I remain bound at NORs as cells enter and exit mitosis.
Nucleolar reformation begins in telophase as processing factors and unprocessed pre-RNA remaining from the previous cell cycle associate with NORs (10 in human), which then cluster into one to five foci. Next, a wide variety of nucleolar components assemble into particles termed prenucleolar bodies that associate with the NORs in a process requiring transcription of the rRNA genes. Normally, nascent transcripts, rather than ribosomal genes, nucleate assembly of the nucleolus in each cell cycle. If antibodies to RNA polymerase I are microinjected into mitotic cells, rRNA transcription is blocked, and nucleoli do not reform in the next G1 phase.
Structure of the Nuclear Envelope
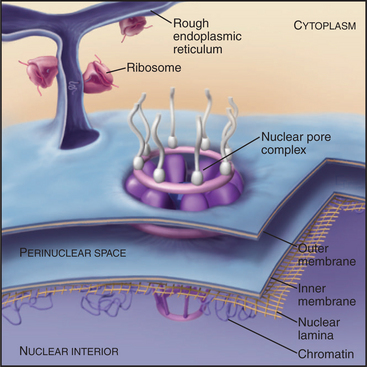
The nuclear envelope provides a selective permeability barrier between the nuclear compartment and the cytoplasm (Fig. 14-5). This barrier ensures that only fully processed mRNAs are delivered to ribosomes for translation into protein. In addition, various chromosomal events, including DNA replication and expression of certain genes, are regulated, at least in part, by changes in the ability of factors to move from the cytoplasm into the nucleus.
Structure and Assembly of the Nuclear Lamina
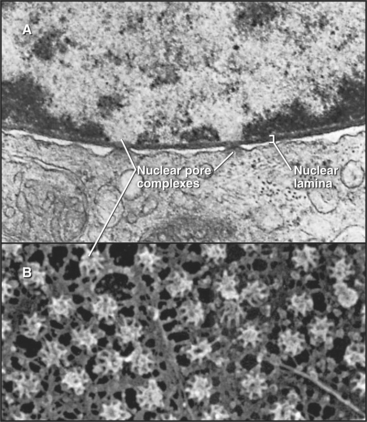
The nuclear lamina is a protein meshwork, typically 20 to 40 nm thick, composed of type V intermediate filament proteins called nuclear lamins (Fig. 14-6). Mammalian lamins are generally divided into two families. Lamin A is encoded by a gene that gives rise to four polypeptides (including lamin C) by alternative splicing (see Fig. 16-6). Members of the lamin B family are the products of two distinct genes.
Lamin gene expression depends on the cell type and stage of development. All nuclei of higher eukaryotes, including early embryos, have a lamina that contains lamin B-family subunits, loss of which is lethal. Lamins A and C typically appear only later in development as cells begin to differentiate. This variation in lamina composition may affect chromosome organization, possibly contributing to different patterns of gene expression.
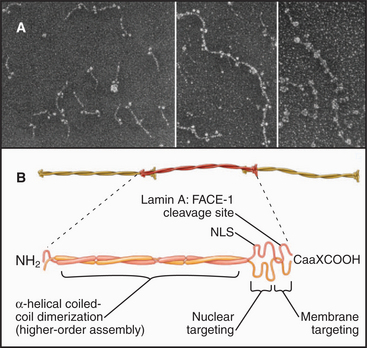
Like other intermediate filament proteins (see Fig. 35-2), nuclear lamins have a central, rod-like domain that is largely a-helical (Fig. 14-7). The basic building block of lamin assembly is an a-helical coiled-coil (see Fig. 3-10) of two identical parallel polypeptides. Two large globular C-terminal domains protrude from one end. Lamin dimers self-associate end to end to form polymers. In some cases, these polymers grow as thick as 10-nm intermediate filaments.
The C-terminal globular domain contains a nuclear localization sequence (see later section) that ensures the rapid import of newly synthesized lamin precursors into the nucleus through nuclear pores. Most lamin subunits acquire a hydrophobic posttranslational modification that targets them to the nuclear membrane. The modification involves the enzymatic addition of a hydrocarbon tail, a C15-isoprenoid group called farnesyl (see Figs. 7-9 and 20-13). The farnesyl group is added to a characteristic amino acid motif called the CaaX box (Ca1a2X, where C refers to cysteine located four amino acids from the carboxyl terminus; a1 refers to any aliphatic amino acid; a2 refers to valine, isoleucine, or leucine; and X refers usually to methionine or serine) at the carboxyl terminus of the protein (Fig. 14-7). This motif was first recognized in the Ras proteins (see Fig. 25-7). Lamin subunits lacking a CaaX box form aggregates in the nuclear interior. Once at the nuclear membrane lamin A is processed by a specialized protease called FACE-1 (farnesylated protein-converting enzyme 1) that clips off the C-terminal 18 amino acids, removing the farnesyl group. The aaX residues are removed from B-type lamins, leaving a protein with the farnesyl group on its carboxyl terminal cysteine.
A diffuse network of lamins is spread throughout the nucleus of most cells, and local concentrations appear at particular times during the cell cycle. Intranuclear lamin A spots are most prominent in G1, whereas those with lamin β are most prominent during S, when they colocalize with sites of DNA replication. Lamin β spots do not colocalize with lamin A spots. The role of these intranuclear concentrations of lamins is unknown, but they might be localized regions of nuclear matrix (see Fig. 13-18) with roles in RNA transcription and DNA replication.
Proteins of the Inner Nuclear Membrane
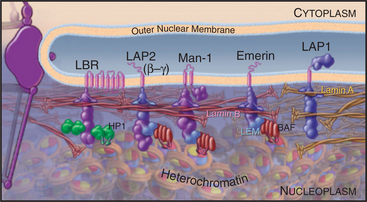
Well-characterized lamin-binding proteins include the lamin β receptor, emerin, and the lamina-associated polypeptides (LAPs; Fig. 14-8). Two unrelated genes—LAP1 and LAP2—produce a number of polypeptides with distinct structural and functional properties as a result of extensive alternative splicing of the primary transcripts.
These four proteins also contribute to organizing chromatin at the nuclear periphery. For example, the lamin β receptor binds heterochromatin protein HP1 (see Fig. 13-9) and could thus link the envelope to condensed heterochromatin. The LEM domain, a degenerate 40-amino-acid motif common to LAP2, emerin, and MAN1, binds to an abundant small protein called barrier to autointegration factor (BAF) that also binds to DNA and has an important role in chromatin organization in both interphase nuclei and mitotic chromosomes. Despite their apparent roles in linking chromosomes to the nuclear envelope, live cell observations have shown that both HP1 and BAF are extremely mobile proteins.
Are these interactions between the chromosomes and the inner nuclear envelope functionally significant? This remains hotly debated, but a number of studies indicate that interactions with LAP2 and lamin A could be important in regulation of the cell cycle by the transcription factor E2F (see Fig. 41-8). Nuclear envelope proteins have been observed to interact with other transcriptional regulators, so (as is discussed later) alterations in gene expression might explain the link between mutations in nuclear envelope proteins and human disease.
Nuclear Envelope Defects Lead to Human Diseases
In 1994, the gene that is mutated in human X-linked Emery-Dreifuss muscular dystrophy was found to encode a protein of the inner nuclear envelope named emerin. This link between the nuclear envelope and human disease was only the tip of an iceberg. Now genetic defects in nuclear envelope proteins are known to cause at least 14 disorders, including muscular dystrophies, lipodystrophies, and neuropathies (diseases of striated muscle, fatty tissue, and the nervous system). The most dramatic of these is Hutchinson-Gilford progeria (Fig. 14-9). Affected individuals are essentially normal at birth, but they appear to age rapidly and die in their early teens of symptoms (including atherosclerosis and heart failure) that are typically associated with extreme age.
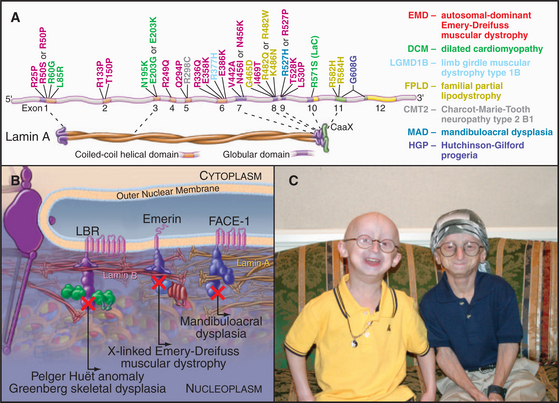
(A, Modified from Mounkes L, Kozlov S, Burke B, Stewart CL: The laminopathies: Nuclear structure meets disease. Curr Opin Genet Dev 13:223–230, 2003. C, Courtesy of the Progeria Research Foundation, Peabody, Massachusetts, http://www.progeriaresearch.org.)
Over 180 mutations scattered throughout the gene encoding both lamin A and C cause over 10 different diseases, collectively termed laminopathies (Fig. 14-9). At least two laminopathies are also linked to mutations in FACE-1, the membrane-associated protease that processes prelamin A. Some of the symptoms of laminopathies can be modeled in the mouse. Loss of lamin A causes disruption of the nuclear envelope and leads to a type of muscular dystrophy. Other mutations in mouse lamin A reproduce aspects of Hutchinson-Gilford pro-geria.
An alternative suggestion is that these mutations cause disease by altering gene expression by compromising interactions between the inner nuclear membrane and chromatin. Cells from patients with Hutchinson-Gilford progeria show signs of aging in culture that are accompanied by dramatic alterations in heterochromatin (see Fig. 13-9), lending support to this model.
Nuclear Pore Complexes
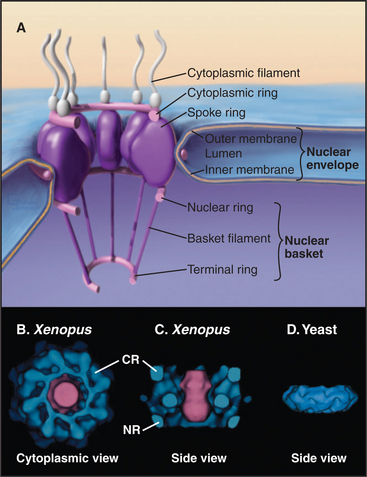
In a typical growing cell, all traffic between the nucleus and cytoplasm passes through 3000 to 5000 channels, called nuclear pore complexes, that bridge both the inner and outer nuclear membranes (Fig. 14-10). Nuclear pore complexes have a central cylindrical core 90 nm long from which filaments project into the cytoplasm and nucleus. The central core consists of a massive multidomain spoke ring with eightfold symmetry surrounded by a luminal ring in the perinuclear space and sandwiched between cytoplasmic and nuclear rings associated with the surfaces of the outer and inner nuclear membranes, respectively. The nuclear ring appears to be anchored to the nuclear lamina. The minimum diameter of the central channel through the spoke ring is 45 to 55 nm.
Vertebrate nuclear pore complexes are large structures with a mass of approximately 90 million to 120 million daltons. Yeast nuclear pores are similar in overall structure but about half the mass. Given their large mass, it was originally believed that vertebrate nuclear pore complexes would contain multiple copies of 100 or more proteins. In fact, the core protein composition of yeast and mammalian pore complexes is remarkably similar. Both are composed of about 30 core proteins known as nucleoporins (Fig. 14-11). These are present in multiples of eight copies. With an average size of more than 100 kD, they can account for the observed mass of the pore. Proteomic analysis of rat and yeast nuclear pore complexes identified 94 and 174 polypeptides, respectively. The additional polypeptides are primarily transport factors and other auxiliary subunits that do not have a key structural role.
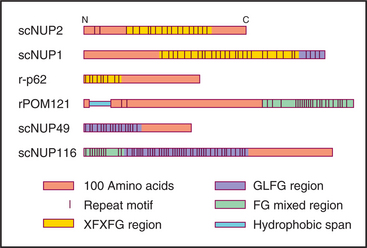
Figure 14-11 sequence organization of several nucleoporins, the structural components of the nuclear pores. Nucleoporins contain combinations of repeated sequences as shown. Letters refer to the amino acids (see Fig. 3-2). They may be somehow involved in helping cargo to traffic through the pores.
Three experiments show that nucleoporins are required to transport proteins into the nucleus. First, antibodies to nucleoporins inhibit transport either when added to isolated nuclei or when injected into live cells. Second, lectins such as wheat germ agglutinin (which binds specifically to sugars attached to many nucleoporins) inhibit transport in similar experiments. Third, nuclear pore complexes assembled in Xenopus egg extracts (see Box 40-3) in the absence of the highly conserved nucleoporin p62, a component of the central region of the cytoplasmic and nuclear faces of the pore complex, appear structurally normal but are inactive in transport.
Traffic between Nucleus and Cytoplasm
The nuclear pore complex is a highly efficient conduit that can allow the passage of up to 1000 macromolecules per second. Traffic heading out of the nucleus includes mRNPs, ribosomal subunits, and transfer RNAs (tRNAs), all of which must be transported to the cytoplasm to function in protein synthesis. Traffic headed into the nucleus includes transcription factors, chromatin components, and ribosomal proteins. Other molecules follow more complex routes. snRNAs are exported to the cytoplasm to acquire essential protein components; they are then reimported into the nucleus, where they undergo further maturation steps before functioning in RNA processing. Individual pores can simultaneously transport components in both directions.
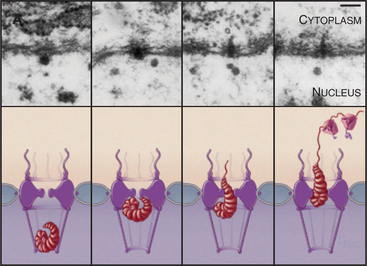
The pore gate opens to a maximum of 30 to 40 nm, but larger particles can squeeze through, provided that they are deformable. This is well documented for export of a well-studied enormous RNA that associates with roughly 500 packaging proteins to make an RNP particle about 50 nm in diameter. The RNP is deformed into a rod-shaped structure as it squeezes through the pore (Fig. 14-12). Rigid particles cannot usually exceed the 30- to 40-nm limit.
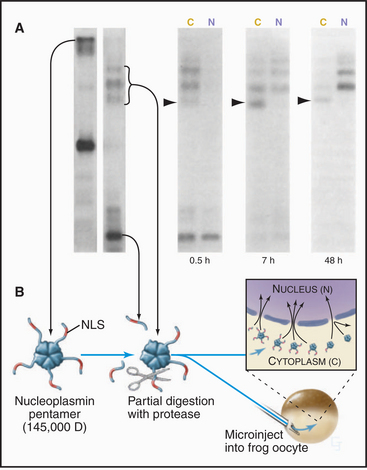
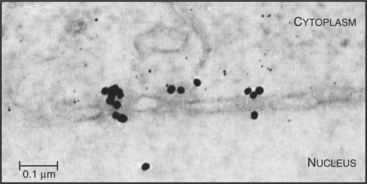
Proteins that are imported into the nucleus bear a nuclear localization sequence (NLS), also called a nuclear localization signal, that is recognized by specific carrier proteins called transport receptors (Figs. 14-13 and 14-14). Known types of NLS vary in complexity. The best studied is a patch of basic amino acids similar to the sequence PKKKRKV (single-letter amino acid code; see Fig. 3-2), first identified on the simian virus 40 (SV40) large T antigen. A point mutation, yielding PKNKRKV, inactivates this sequence as a signal for nuclear transport. A related type of bipartite NLS features two smaller patches of basic residues separated by a variable spacer (KRPAATKKAGQAKKKK [critical residues are underlined]). These two types of sequences are referred to as basic NLSs. Basic NLSs function autonomously and can direct the migration of a wide range of molecules into the nucleus in vivo. In one extreme example, when coated with nucleoplasmin, a protein with a bipartite basic NLS, colloidal gold particles up to 23 nm in diameter are transported through nuclear pores (Fig. 14-15). An alternative type of NLS is not basic but instead is rich in glycine. Proteins with this NLS are imported by a similar mechanism (see later) but are recognized by a different transport receptor.
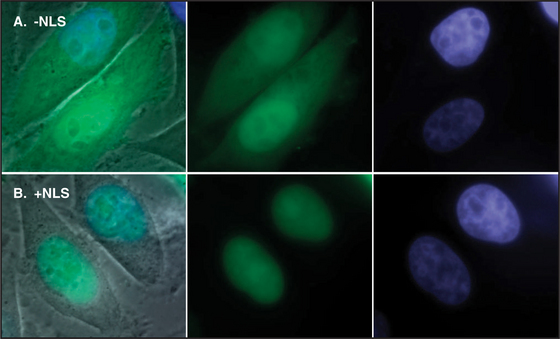
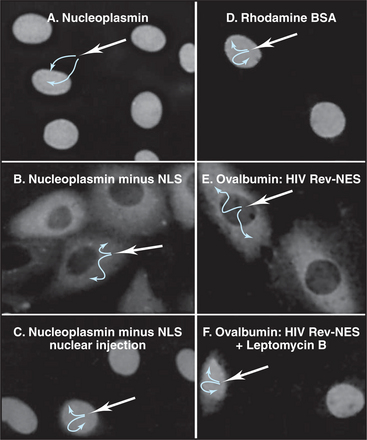
Figure 14-14 ICAD protein (the inhibitor of CAD nuclease; see Chapter 46) was fused to the green fluorescent protein (GFP) and expressed in cultured cells. A, A mutant form of the ICAD: GFP fusion protein lacking the ICAD nuclear localization se-quence (NLS) accumulates randomly throughout the cell. B, The intact ICAD: GFP fusion protein with NLS accumulates quantitatively in the nucleus.
(Courtesy of K. Samejima, University of Edinburgh, Scotland.)
Many proteins exported from the nucleus bear a nuclear export sequence (NES) that is recognized by carriers related to those used for nuclear import (Fig. 14-16). Like import signals, these signals vary in size and complexity. The human immunodeficiency virus I Rev protein provides one example of a leucine-rich sequence (LQLPPLERLTL) that is recognized by the carrier CRM1. Certain RNA sequences or structures may also serve as NESs.
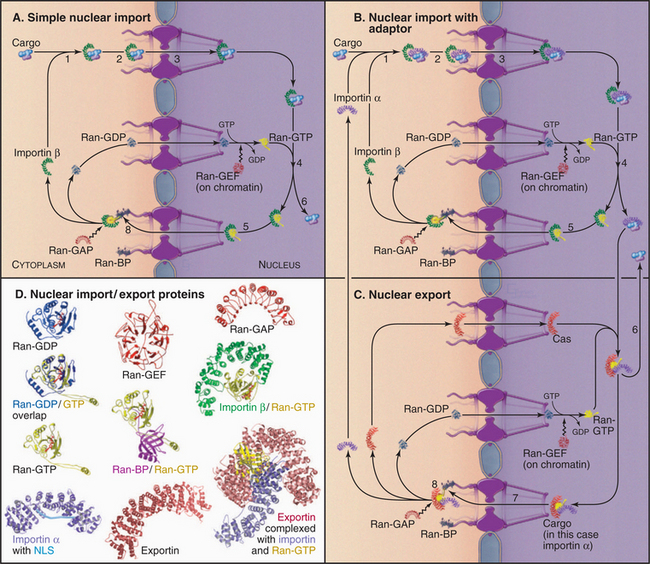
The following is a brief thumbnail of protein import into the nucleus (Fig. 14-17). A protein with an NLS (known as cargo) binds to an import receptor either by itself or in combination with an adapter molecule, forming a complex, which then passes through pores into the nucleus. There, the cargo and adapter (if used) are displaced from the import receptor. The adapter then releases its cargo and is transported back to the cytoplasm as the cargo of an export receptor. Import receptors also shuttle back through pores, where they can meet more cargo or cargo/adapter complexes. Molecules exported from the nucleus use a variation of this cycle, being picked up by the transport machinery in the nucleus and discharged in the cytoplasm.
The key to this system is that it is vectorial: Nuclear components are transported into the nucleus while components that function in the cytoplasm are transported out. This means that each carrier picks up its cargo on one side of the nuclear envelope and deposits it on the other. This directionality is regulated by a simple yet elegant system involving Ran, a small guanine triphosphatase (GTPase [see Figs. 4-6 and 4-7 for background material on GTPases]), and associated factors.
Components of Nuclear Import and Export
The nuclear import and export system involves many components, but the general principles of its operation are simple. To understand how it works, this section first introduces several of the components (Table 14-2) and then describes one transport event in detail.
Table 14-2 SEVERAL KEY PROTEINS INVOLVED IN NUCLEAR TRAFFICKING
| Adapters | |
| Importin α* | An import adapter, importin α interacts with importin β during nuclear transport and binds basic NLSs. Humans have at least six distinct importin α genes, expressed in a tissue-specific manner. |
| Snurportin 1 | This adapter binds the trimethyl G cap structure on snRNPs during import. It also interacts with importin β during nuclear transport. |
| hnRNP A1 | This protein has a major role in the export of mRNA from the nucleus. The molecule is extremely abundant (108 copies/cell) and active (105 copies/min shuttling between the nucleus and cytoplasm). |
| Importin β—Related Molecules Involved in Import* | |
| Importin β | A founder member of the large family of nuclear trafficking receptors, importin β binds various adapters and interacts with nucleoporins during import. Ran-GTP regulates binding to adapters. |
| Transportin | Transportin imports mRNA-binding proteins into the nucleus. |
| Importin β—Related Molecules Involved in Export | |
| CAS | CAS recycles importin α and snurportin 1 to the cytoplasm. |
| CRM1 | CRM1 exports proteins with leucine-rich NESs and also U snRNAs. It is a target of the fungal toxin leptomycin B. |
| Exportin-t | This protein is involved in tRNA export. |
| Directionality Factors | |
| Ran | A small Ras-family GTPase, Ran binds importin-related nuclear trafficking receptors, as well as a number of regulatory proteins. |
| Ran-GAP1 | This protein stimulates GTP hydrolysis by Ran in cytoplasm (GAP = GTPase-activating protein). |
| RCC1 | RCC1 is the nuclear GTP exchange factor (GEF) for Ran. It stimulates release of GDP from Ran. |
| Ran-BP1 | Ran-BP1 is a cytoplasmic protein that binds Ran-GTP and acts with Ran-GAP1 to stimulate GTP hydrolysis by Ran. |
| Ran-BP2 | A major component of the cytoplasmic filaments of the nuclear pore, Ran-BP2 has four Ran-binding domains. It also binds Ran-GAP1 and may locally convert cytoplasmic Ran-GTP to Ran-GDP so that it can return to the nucleus. |
| NTF2 | NTF2 binds RanGDP and promotes its recycling back to the nucleus. |
* The importin molecules (humans have 14 genes) were discovered independently and termed karyopherins.
Adapters
Adapters bind to the NLS or NES sequences on some cargo molecules and also to particular regions on receptors. The best-known adapter is importin α, which is responsible for recognition of small basic NLS sequences and works together with the transport receptor importin β (see later) in nuclear transport. Importin α consists of a highly flexible N-terminal NLS-like importin β-binding domain followed by 10 repeats of a helical motif (the Armadillo repeat [Fig. 14-17D]) that give the structured portion of the molecule a slug-like shape. The importin β-binding motif can bind either the NLS-binding region on importin β or the NLS-binding domain on importin α itself (the “belly” of the slug). The latter provides an autoinhibitory mechanism that is thought to be important in regulating the release of cargo in the nucleus at the end of an import cycle. Binding to importin β uncovers the NLS binding site on importin α so that it can bind cargo more efficiently.
Receptors
With the exception of mRNP export from the nucleus (which uses special transport factors), all nuclear trafficking receptors are related to importin β, the import receptor for proteins bearing a basic NLS. At least 20 members of the importin β family are known in vertebrates (14 in yeast). These proteins are also called karyopherins. Some of these function in nuclear import, whereas others function in export. Importin β consists entirely of 19 copies of a helical protein interaction motif called a HEAT repeat, giving the protein the shape of a snail-like superhelix with the potential to interact with a large number of protein ligands. All importin β family members have a binding site for the Ran GTPase that encompasses several HEAT repeats at the N-terminus (Fig. 14-17D). Importin β binds many NLSs directly but also interacts with other cargoes via the importin α adapter. Other importin β HEAT repeats bind to FG repeats of the nucleoporins.
Directionality/Recycling Factors
Like other small GTPases, Ran has low intrinsic GTPase activity, but interactions with binding proteins (Ran-BP1 or Ran-BP2) and a GTPase-activating protein called Ran-GAP1 stimulate GTP hydrolysis. Ran-BP1 is anchored in the cytoplasm. Ran-BP2 is a component of the fibers projecting from the nuclear pore into the cytoplasm. This huge (>350 kD) protein can bind up to four Ran molecules as well as Ran-GAP1 and may provide a structural scaffold for the conversion of Ran-GTP into Ran-GDP at the surface of the pore. Because Ran-BP1 and Ran-BP2 are both anchored in the cytoplasm, Ran-GTP is efficiently converted to Ran-GDP only in the cytoplasm, yielding a nuclear/cytoplasmic ratio of Ran-GTP of ˜200:1.
Ran-GDP must reenter the nucleus to be recharged with GTP. Efficient Ran-GDP transport into the nucleus requires nuclear transport factor 2 (NTF2). Back in the nucleus, Ran must release its bound GDP to acquire GTP. GDP dissociation is slow but is stimulated by a guanine nucleotide exchange factor (GEF). This protein, called regulator of chromosome condensation 1 (RCC1), is tightly associated with chromatin through-out the cell cycle. This allows nuclear import to resume immediately after the nuclear envelope reforms at the end of mitosis. Since Ran is involved in essential every nuclear trafficking event, the flux of this small protein across the nuclear envelope is enormous—several million molecules per minute in cultured cells.
Description of a Single Import Cycle in Detail
Consider the import into the nucleus of a typical protein (Fig. 14-17):
Regulation of Transport across the Nuclear Envelope
Nuclear trafficking is most commonly regulated by phosphorylation near the NLS on the cargo. Phosphorylation adjacent to a basic NLS inhibits nuclear import. This provides a mechanism to regulate the ability of a particular cargo to enter the nucleus in response to cell cycle (see Fig. 43-6) or other cues that can be coupled to specific protein kinase activation.
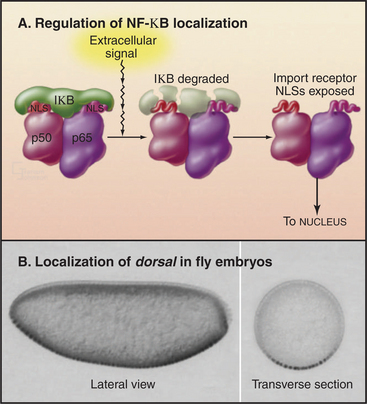
Traffic across the nuclear envelope may be also regulated by masking or unmasking nuclear localization sequences. A “nuclear” protein with a masked NLS is trapped in the cytoplasm. A good example is the regulation of transcription factor NF-kB by IkB (Fig. 14-18). IkB binds to NF-kB and covers up its NLS. Because IkB also has a nuclear export signal, the NF-kB: IkB complex is entirely cytoplasmic. Following an appropriate signal (see Fig. 15-22C), IkB is degraded. This uncovers the NLS on NF-kB, allowing it to enter the nucleus.
Disorders Associated with Defective Nuclear Trafficking
In many instances, protein function appears to be regulated by adjusting its location in the cell, and nuclear transport is one mechanism controlling localization. Thus, a myriad of examples undoubtedly exist in which disruption of transport leads to disease. This area has yet to be explored systematically, but in one interesting example, human sex determination is disrupted by mutations of a NLS on the SRY transcription factor, a master regulator of sex determination. These NLS mutants apparently disrupt the accumulation of SRY in the nucleus at a critical stage during development, causing individuals with a 46XY karyotype (normal male) to develop as females.
Azuma Y, Dasso M. The role of Ran in nuclear function. Curr Opin Cell Biol. 2000;12:302-307.
Harel A, Forbes D. Importin β Conducting a much larger cellular symphony. Mol Cell. 2004;16:319-330.
Hetzer MW, Walther TC, Mattaj IW. Pushing the envelope: Structure, function, and dynamics of the nuclear periphery. Annu Rev Cell Dev Biol. 2005;21:347-380.
Hood JK, Silver PA. In or out? Regulating nuclear transport. Curr Opin Cell Biol. 1999;11:241-247.
Hutchison CJ, Alvarez-Reyes M, Vaughan OA. Lamins in disease: Why do ubiquitously expressed nuclear envelope proteins give rise to tissue-specific disease phenotypes? J Cell Sci. 2001;114:9-19.
Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547-553.
Lamond AI, Spector DL. Nuclear speckles: A model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605-612.
Lewis JD, Tollervey D. Like attracts like: Getting RNA processing together in the nucleus. Science. 2000;288:1385-1389.
Matera AG. Nuclear bodies: Multifaceted subdomains of the interchromatin space. Trends Cell Biol. 1999;9:302-309.
Mattaj IW, Englmeier L. Nucleocytoplasmic transport: The soluble phase. Annu Rev Biochem. 1998;67:265-306.
Mounkes L, Kozlov S, Burke B, Stewart CL. The laminopathies: Nuclear structure meets disease. Curr Opin Genet Dev. 2003;13:223-230.
Nigg EA. Nucleocytoplasmic transport: Signals, mechanisms and regulation. Nature. 1997;386:779-787.
Pemberton LF, Blobel G, Rosenblum JS. Transport routes through the nuclear pore complex. Curr Op Cell Biol. 1998;10:392-399.
Spector DL. The dynamics of chromosome organization and gene regulation. Annu Rev Biochem. 2003;72:573-608.
Stuurman N, Heins S, Aebi U. Nuclear lamins: Their structure, assembly, and interactions. J Struct Biol. 1998;122:42-66.
Suntharalingham M, Wente SR. Peering through the pore: Nuclear pore complex structure, assembly and function. Dev Cell. 2003;4:775-789.