Molecular Pathology
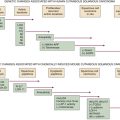
Hereditary Prostate Cancer
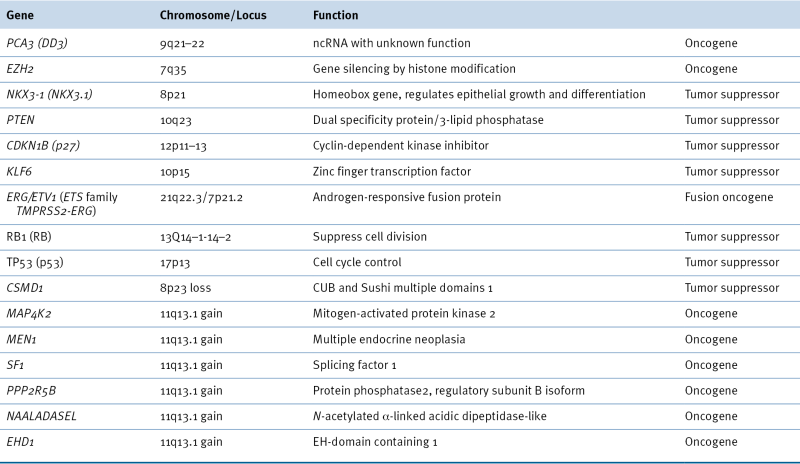
Table 38-1
Oncogenes and Tumor Suppressors Implicated in Prostate Cancer
Gene Mutation
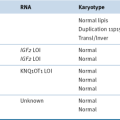
DNA Copy Number Variation
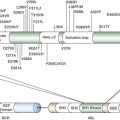
Gene Fusion
WNT Signaling and β-Catenin
MicroRNA and Other Noncoding RNA in Prostate Cancer
MicroRNA and Prostate Cancer
Expression Signature of miRNA in Prostate Cancer
miRNA and the AR Signaling Pathway
miRNA as Biomarkers for Prostate Cancer Diagnosis
lncRNA and Prostate Cancer
Epigenetic Alterations in Prostate Cancer
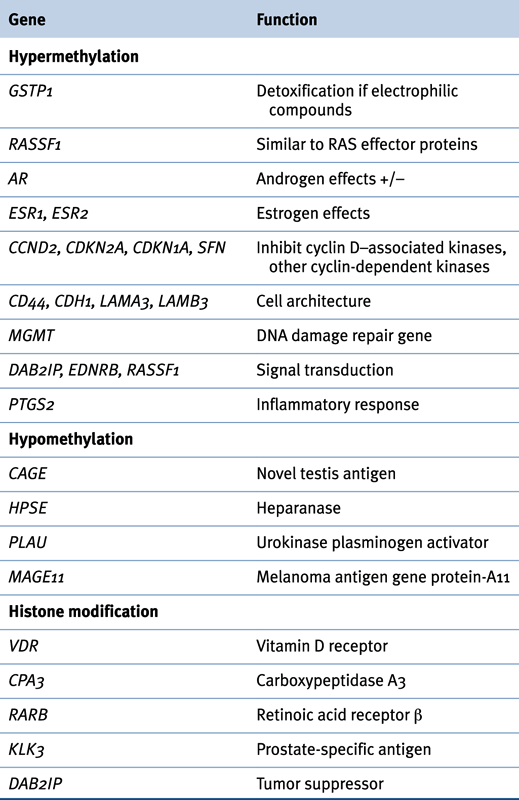
Hypermethylation
DNA Damage-Repair Genes
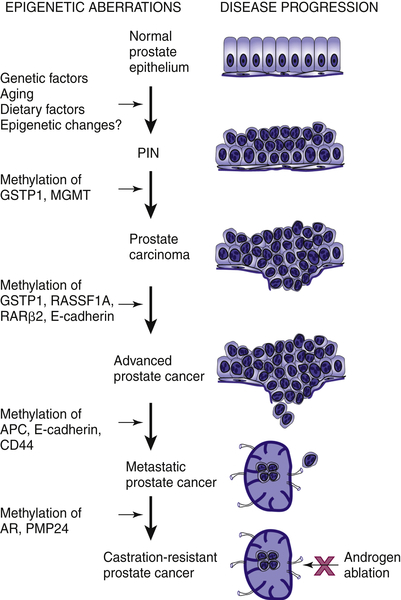
Table 38-2
Epigenetic Changes in Prostate Cancer
Hormone Receptors
Cell Cycle Control Genes
Tumor Invasion and Tumor Architecture Genes
Other Putative Tumor Suppressor Genes
Hypomethylation
Histone Modification
Polycomb Group Transcriptional Repression
PSA
Aberrant Translational Control in Prostate Cancer Etiology and Progression
Future Directions
1. Cancer statistics, 2012 . CA Cancer J Clin . 2012 ; 62 : 10 – 29 .
2. International trends in prostate-cancer mortality: the decrease is continuing and spreading . Cancer Causes Control . 2004 ; 15 : 237 – 241 .
3. Mortality results from a randomized prostate-cancer screening trial . N Engl J Med . 2009 ; 360 : 1310 – 1319 .
4. Screening and prostate-cancer mortality in a randomized European study . N Engl J Med . 2009 ; 360 : 1320 – 1328 .
5. Heterogeneity of Gleason grade in multifocal adenocarcinoma of the prostate . Cancer . 2004 ; 100 : 2362 – 2366 .
6. A systematic review and meta-analysis of familial prostate cancer risk . 29BJU Int . 2003 ; 91 : 789 – 794 .
7. The mutational landscape of lethal castration-resistant prostate cancer . Nature . 2012 ; 487 : 239 – 243 .
8. Integrative genomic profiling of human prostate cancer . Cancer Cell . 2010 ; 18 : 11 – 22 .
9. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer . Cancer Res . 2006 ; 66 : 3396 – 3400 .
10. miRNAs as biomarkers in prostate cancer . Clin Transl Oncol . 2012 ; 14 : 803 – 811 .
11. miR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth . Cancer Res . 2009 ; 69 : 7165 – 7169 .
12. Microfluidic-based multiplex qRT-PCR identifies diagnostic and prognostic microRNA signatures in the sera of prostate cancer patients . Cancer Res . 2011 ; 71 : 550 – 560 .
13. microRNA biomarkers in body fluids of prostate cancer patients . Methods . 2013 ; 59 : 132 – 137 .
14. Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer . Oncogene . 2004 ; 23 : 6684 – 6692 .
15. The prostate cancer-up-regulated long noncoding RNA PlncRNA-1 modulates apoptosis and proliferation through reciprocal regulation of androgen receptor . Urol Oncol . 2013 ; 31 ( 7 ) : 1117 – 1123 .
16. Up-regulation of Wnt-1 and beta-catenin production in patients with advanced metastatic prostate carcinoma: potential pathogenetic and prognostic implications . Cancer . 2004 ; 101 : 1345 – 1356 .
17. Global reactivation of epigenetically silenced genes in prostate cancer . Cancer Prev Res (Phila) . 2010 ; 3 : 1084 – 1092 .
18. Discovery of novel hypermethylated genes in prostate cancer using genomic CpG island microarrays . PLoS One . 2009 ; 4 : e4830 .
19. Genome-wide methylation analysis identifies involvement of TNF-alpha mediated cancer pathways in prostate cancer . Cancer Lett . 2011 ; 302 : 47 – 53 .
19a. Constitutively active Akt1 expression in mouse pancreas requires S6 kinase 1 for insulinoma formation . J Clin Invest . 2008 ; 118 : 3629 – 3638 . doi: 10.1172/JCI35237 .
19b. Genetic Dissection of the Oncogenic mTOR Pathway Reveals Druggable Addiction to Translational Control via 4EBP-eIF4E . Cancer Cell . 2010 ; 17 : 249 – 261 . doi: 10.1016/j.ccr.2010.01.021 .
19c. Janes MR, et al. Effective and selective targeting of leukemia cells using a TORC1/2 kinase inhibitor. Nat Med. 16, 205–213, doi:nm.2091 [pii]
19d. Gene amplifications associated with the development of hormone-resistant prostate cancer . Clin Cancer Res . 2003 ; 9 : 5271 – 5281 .
19e. Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer cell 18, 11–22, doi:S1535-6108(10)00238-2 [pii].10.1016/j.ccr.2010.05.026
19f. Control of p70 s6 kinase by kinase activity of FRAP in vivo . Nature . 1995 ; 377 : 441 – 446 . doi: 10.1038/377441a0 .
19g. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway . Genes & development . 1998 ; 12 : 502 – 513 .
19h. An essential E box in the promoter of the gene encoding the mRNA cap-binding protein (eukaryotic initiation factor 4E) is a target for activation by c-myc . Mol Cell Biol . 1996 ; 16 : 4754 – 4764 .
19i. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis . Nat Med . 2004 ; 10 ( 5 ) : 484 – 486 .
19j. Targeting synthetic lethal interactions between Myc and the eIF4F complex impedes tumorigenesis . Cell reports . 2012 ; 1 : 325 – 333 . doi: 10.1016/j.celrep.2012.02.010 .
19k. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2 . The EMBO journal . 1997 ; 16 : 1909 – 1920 . doi: 10.1093/emboj/16.8.1909 .
19l. The translational landscape of mTOR signalling steers cancer initiation and metastasis . Nature . 2012 doi: 10.1038/nature10912 .
20. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression . Proc Natl Acad Sci U S A . 2010 ; 107 : 14134 – 14139 .
21. The prostate cancer conundrum revisited: treatment changes and prostate cancer mortality declines . Cancer . 2012 ; 118 : 5955 – 5963 .
22. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5’-cap structure . J Biol Chem . 1997 ; 272 ( 35 ) : 21677 – 21680 .
23. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A . 1999 ; 274 ( 18 ) : 12236 – 12244 .
24. Lahat G, et al. Vimentin is a novel anti-cancer therapeutic target; insights from in vitro and in vivo mice xenograft studies. PloS one 5. e10105, 10.1371/journal.pone.0010105
25. Liu C, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nature. medicine 17. 211–215, doi:nm.2284 [pii]10.1038/nm.2284
26. The role of metastasis-associated protein 1 in prostate cancer progression . Cancer research . 2004 ; 64 : 825 – 829 .
27. Yoo YG, Kong G, Lee MO. Metastasis-associated protein 1 enhances stability of hypoxia-inducible factor-1alpha protein by recruiting histone deacetylase 1. The EMBO journal 25. 1231–1241, doi:7601025 [pii]
28. Evdokimova V, et al. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell 15. 402–415, doi:S1535-6108(09)00086-5 [pii]
29. Targeting the eIF4F translation initiation complex for cancer therapy . Cell Cycle . 2008 ; 7 : 2466 – 2471 doi:6464 [pii] .
30. Translational control in cancer . Nature reviews. Cancer . 2010 ; 10 : 254 – 266 . doi: 10.1038/nrc2824 .
31. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression . Proc Natl Acad Sci U S A . 2010 ; 107 ( 32 ) : 14134 – 14139 .
32. eIF4E activation is commonly elevated in advanced human prostate cancers and significantly related to reduced patient survival . Cancer research . 2009 ; 69 : 3866 – 3873 doi:0008-5472.CAN-08-3472[pii]10.1158/0008-5472.CAN-08-3472 .
33. An oncogenic role for the phosphorylated h-subunit of human translation initiation factor eIF3 . The Journal of biological chemistry . 2008 ; 283 : 24047 – 24060 . doi: 10.1074/jbc.M800956200 .
34. Angiogenin-stimulated rRNA transcription is essential for initiation and survival of AKT-induced prostate intraepithelial neoplasia . Molecular cancer research: MCR . 2009 ; 7 : 415 – 424 . doi: 10.1158/1541-7786.MCR-08-0137 .
35. Overexpression of ribosomal RNA in prostate cancer is common but not linked to rDNA promoter hypomethylation . Oncogene . 2012 ; 32 : 1254 – 1263 . doi: 10.1038/onc.2011.319 .
36. DKC1 overexpression associated with prostate cancer progression . Br J Cancer . 2009 ; 101 : 1410 – 1416 . doi: 10.1038/sj.bjc.6605299 .
37. Translational control in cancer etiology . Cold Spring Harb Perspect Biol . 2013 ; 5 : a012336 .
38. Emerging therapeutics targeting mRNA translation . Cold Spring Harb Perspect Biol . 2012 ; 4 : a012377 .