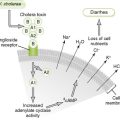
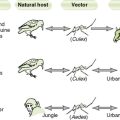
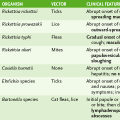
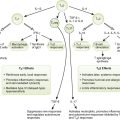
Laboratory Tests for Diagnosis
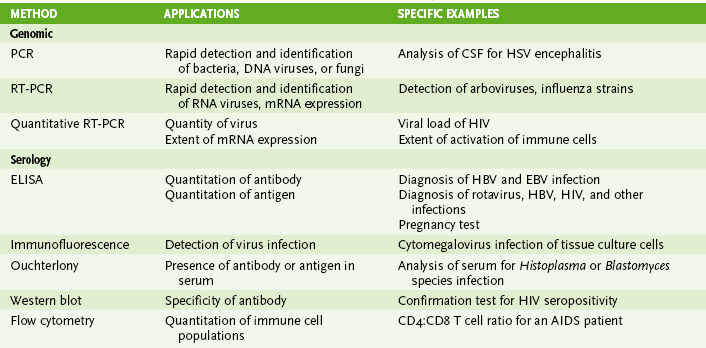
I Laboratory Assays for Detecting Nucleic Acids (Table 5-1)
• Detection of RNA or DNA is rapid and sensitive and can detect microbes that are too virulent or not readily grown in the laboratory.
• Methods depend on hybridization of a probe (primer) sequence with a complementary sequence in the sample.
• Probes are either radioactive or chemically labeled to allow detection on hybridization with sample.
A Southern blotting for DNA and Northern blotting for RNA detect electrophoretically separated genome sequences.
B In situ hybridization detects viral DNA or RNA within infected cells.
C Polymerase chain reaction (PCR) (DNA), reverse transcriptase (RT)-PCR (RNA), and related technologies permit detection, identification, and amplification of specific nucleic acid sequences.
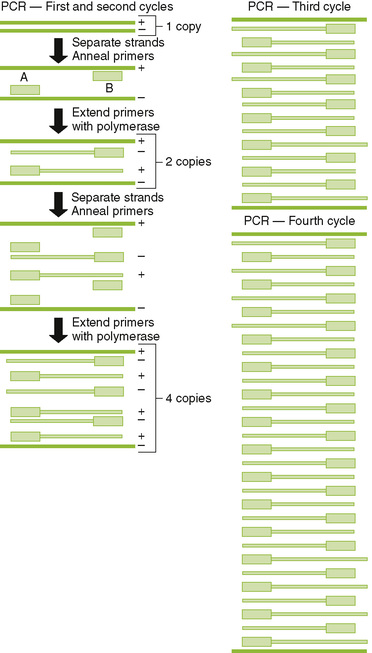
1. PCR: heat-stabile DNA polymerase amplifies DNA between sequence specific primers (Fig. 5-1)
2. RT-PCR: RT makes a complementary DNA copy of RNA, which is then amplified by PCR using sequence specific primers.
• Detects RNA virus genomes, such as HIV
• Detects messenger RNA for cytokine genes activated in an immune response
3. Quantitative PCR (qPCR; real-time PCR): rate of production of DNA during PCR reaction determines concentration of DNA in sample.
• qPCR after reverse transcription of HIV genome can be used to quantitate the serum concentration of virus (genome load of infection).
4. Other DNA and RNA detection methods include branched-chain DNA assay and antibody capture solution hybridization assay.
II Immunologic Assays (seeTable 5-1)
• Assays can be used to detect immune responses to infection, or specific antibody can be used to detect soluble and cell-associated antigen.
A Antibody-antigen binding (see alsoChapter 3, section IV)
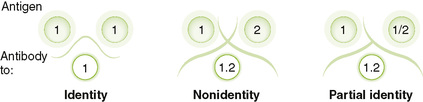
1. Precipitin reactions: Ouchterlony immunodiffusion distinguishes identical, similar, and different antigens based on the precipitin line formed by antigen-antibody precipitation (Fig. 5-2)
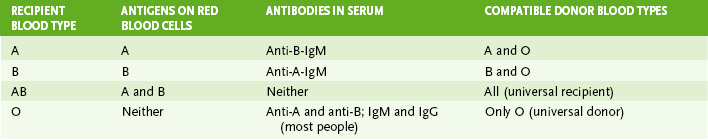
2. Agglutination reactions include hemagglutination, passive agglutination, Coombs test, and ABO blood typing (Table 5-2).
3. Antibody-antigen binding is detected by a probe such as a fluorescent marker, radiolabel, or enzyme (e.g., horseradish peroxidase, alkaline phosphatase, and β-galactosidase) that produces a colored product on addition of substrate.
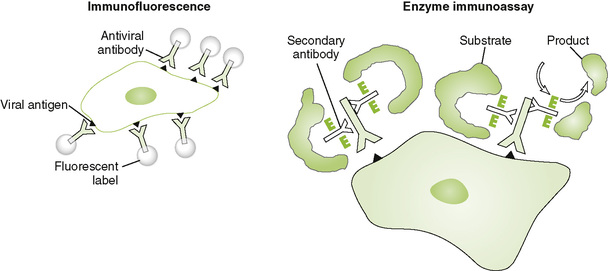
B Immunofluorescence (IF) and enzyme immunoassay (EIA) detect proteins expressed on the surface or inside of cells (Fig. 5-3).
C Direct versus indirect assays
1. For direct assays, antibody is covalently linked to the probe.
2. For indirect assays, the probe is linked to a secondary antibody that binds to the primary antiviral antibody after it interacts with viral antigen.
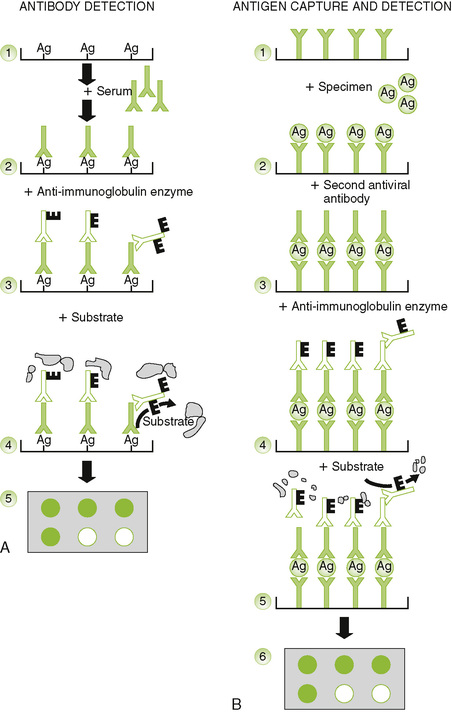
D Enzyme-linked immunosorbent assay (ELISA) and radioimmunoassay (RIA) can be used to detect and quantitate antigen or antibody.
• For analysis of antigen: antibody is attached to plate and captures antigen, and an enzyme-linked antibody is used to detect and quantitate the antigen.
• For analysis of antibody: antigen is attached to plate, antibody binds, and an enzyme-linked anti-antibody is used to detect and quantitate the antigen.
2. RIA: direct or competitive methods are used to detect and quantitate antigen.
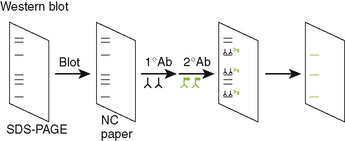
E Western blot can determine true-positive ELISA reactions by showing the specific proteins recognized by patient sera.
1. Western Blot analysis uses antibody to identify proteins that were blotted onto special paper membranes after molecular size separation by sodium dodecyl sulfate polyacrylamide gel electrophoresis (Fig. 5-5).
2. True-positive reactions in ELISA can be confirmed by serum recognition of multiple proteins in the Western blot (confirmation test for HIV).
F Antibody inhibitory tests: hemagglutination inhibition, virus or toxin neutralization
III Serology (History of the Infection) (Box 5-1)
A Serologic testing can determine the type and titer of antiviral antibodies in serum and the identity of viral antigens.
1. Titer is defined as the lowest dilution that still gives a positive test result in a standardized assay.
2. A fourfold difference in antibody titer is required to indicate a significant response to antigen (seroconversion). (A fourfold difference means two tubes in a dilution scheme.)
3. A fourfold increase in antibody titer between acute and convalescent sera is necessary to indicate specific response.
4. Detection of specific immunoglobulin M (IgM) indicates recent exposure to antigen.
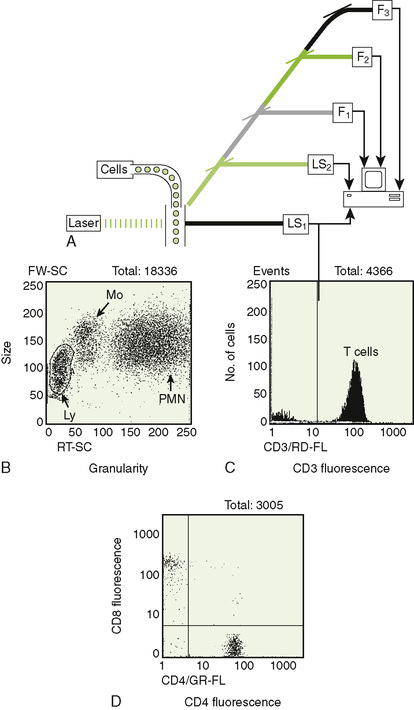
• Laser is used to rapidly evaluate size, granularity, and fluorescence properties of large numbers of cells (Fig. 5-6).
• A fluorescence-activated cell sorter (FACS) is a flow cytometer that can analyze and separate cells based on their properties as they flow through the instrument.
A Light scatter measurements can distinguish lymphocytes, macrophages, and granulocytes.
B DNA-binding fluorescent dyes can be used to evaluate cell cycle.
1. Cells in G0,G1 phase have fluorescence equivalent to 2N (N = normal chromosome number).
2. Cells in G2 phase or mitosis have twice the fluorescence of G0,G1 phase cells.
C Immunofluorescence measurements indicate the phenotype of the cell.
1. Multiple antibodies marked with different fluorescent colors can be analyzed simultaneously.
2. Cells can be identified and quantitated by analysis of multiple cell surface markers (e.g., helper T cell expresses CD3, CD4).
3. Analysis can be presented as a histogram or a two-dimensional dot plot (see Fig. 5-6B to D).