Techniques to Modify the Mouse Genome
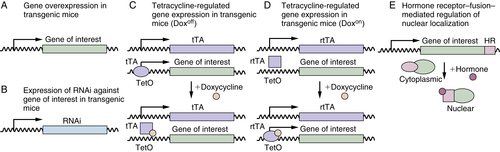
Transgenic Mice
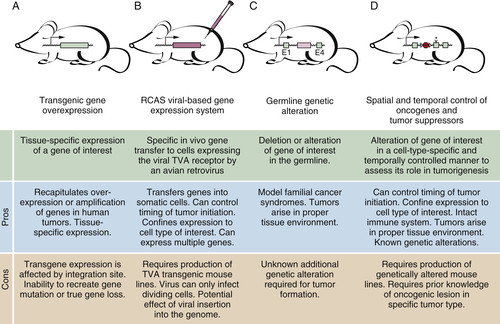
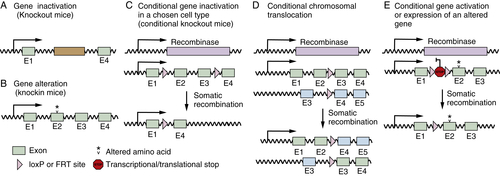
Gene-Targeted Mice
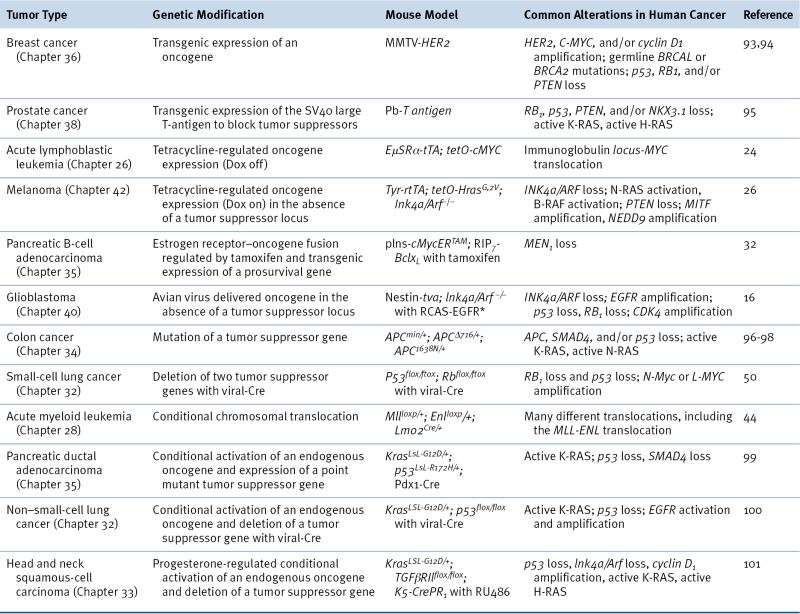
Applications of Mouse Models to Cancer Biology
Cross-Species Comparisons
Table 9-1
Examples of Genetically Modified Mouse Models of Cancer
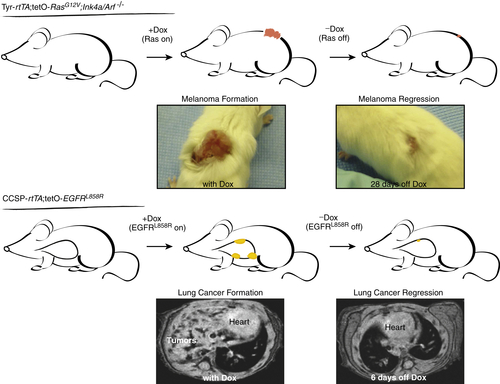
Oncogene Addiction
Oncogene Cooperation and Codependence
Future Directions of Cancer Models
In Vivo Screens
Validation of Pharmaceutical Targets and Preclinical Trials
Biomarkers for Early Tumor Detection
Identification of the Cell of Origin
Recruitment and Function of Immune, Vascular, and Stromal Cells in the Tumor Environment
Conclusions
1. Characterization of a 54K Dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells . Cell . 1979 ; 17 : 43 – 52 .
2. T antigen is bound to a host protein in SV40-transformed cells . Nature . 1979 ; 278 : 261 – 263 .
3. DNA and RNA from uninfected vertebrate cells contain nucleotide sequences related to the putative transforming gene of avian myelocytomatosis virus . J Virol . 1979 ; 31 : 514 – 521 .
4. DNA related to the transforming gene(s) of avian sarcoma viruses is present in normal avian DNA . Nature . 1976 ; 260 : 170 – 173 .
5. Cancer modeling in the modern era: progress and challenges . Cell . 2002 ; 108 : 135 – 144 .
6. Forty years of cancer modelling in the mouse . Eur J Cancer . 2004 ; 40 : 1974 – 1980 .
7. The mighty mouse: genetically engineered mouse models in cancer drug development . Nat Rev Drug Discov . 2006 ; 5 : 741 – 754 .
8. Using genetically engineered mouse models of cancer to aid drug development: an industry perspective . Clin Cancer Res . 2006 ; 12 : 5312 – 5328 .
9. Making waves in cancer research: new models in the zebrafish . Biotechniques . 2005 ; 39 : 227 – 237 .
10. Genome-wide RNAi screens in Caenorhabditis elegans: impact on cancer research . Oncogene . 2004 ; 23 : 8340 – 8345 .
11. Drosophila models for cancer research . Curr Opin Genet Dev . 2006 ; 16 : 10 – 16 .
12. A genetic screen in Drosophila for metastatic behavior . Science . 2003 ; 302 : 1227 – 1231 .
13. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice . Nature . 1985 ; 318 : 533 – 538 .
14. Heritable formation of pancreatic beta-cell tumours in transgenic mice expressing recombinant insulin/simian virus 40 oncogenes . Nature . 1985 ; 315 : 115 – 122 .
15. Transgenic mice harboring SV40 T-antigen genes develop characteristic brain tumors . Cell . 1984 ; 37 : 367 – 379 .
16. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice . Genes Dev . 1998 ; 12 : 3675 – 3685 .
17. Tissue-specific and reversible RNA interference in transgenic mice . Nat Genet . 2007 ; 39 : 914 – 921 .
18. A rapid and scalable system for studying gene function in mice using conditional RNA interference . Cell . 2011 ; 145 : 145 – 158 .
19. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters . Proc Natl Acad Sci U S A . 1992 ; 89 : 5547 – 5551 .
20. Transcriptional activation by tetracyclines in mammalian cells . Science . 1995 ; 268 : 1766 – 1769 .
21. Temporal control of gene expression in transgenic mice by a tetracycline-responsive promoter . Proc Natl Acad Sci U S A . 1994 ; 91 : 93029306 .
22. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes . Genes Dev . 2001 ; 15 : 3249 – 3262 .
23. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies . Cancer Cell . 2006 ; 9 : 485 – 495 .
24. Reversible tumorigenesis by MYC in hematopoietic lineages . Mol Cell . 1999 ; 4 : 199 – 207 .
25. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer . Nature . 2004 ; 431 : 1112 – 1117 .
26. Essential role for oncogenic Ras in tumour maintenance . Nature . 1999 ; 400 : 468 – 472 .
27. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors . Genes Dev . 2006 ; 20 : 1496 – 1510 .
28. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis . Cancer Cell . 2002 ; 2 : 451 – 461 .
29. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins . Nucleic Acids Res . 1995 ; 23 : 1686 – 1690 .
30. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases . Nucleic Acids Res . 1999 ; 27 : 4324 – 4327 .
31. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression . Nature . 2006 ; 443 : 214 – 217 .
32. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression . Cell . 2002 ; 109 : 321 – 334 .
33. Bcl-xL gain of function and p19 ARF loss of function cooperate oncogenically with Myc in vivo by distinct mechanisms . Cancer Cell . 2006 ; 10 : 113 – 120 .
34. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century . Nat Rev Genet . 2005 ; 6 : 507 – 512 .
35. Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse . Nature . 1992 ; 359 : 295 – 300 .
36. Tumour predisposition in mice heterozygous for a targeted mutation in Nf1 . Nat Genet . 1994 ; 7 : 353 – 361 .
37. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours . Nature . 1992 ; 356 : 215 – 221 .
38. Role of the INK4a locus in tumor suppression and cell mortality . Cell . 1996 ; 85 : 27 – 37 .
39. Type 1 neurofibromatosis gene: identification of a large transcript disrupted in three NF1 patients . Science . 1990 ; 249 : 181 – 186 .
40. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms . Science . 1990 ; 250 : 1233 – 1238 .
41. Germ-line transmission of a mutated p53 gene in a cancer-prone family with Li-Fraumeni syndrome . Nature . 1990 ; 348 : 747 – 749 .
42. Identification and characterization of the familial adenomatous polyposis coli gene . Cell . 1991 ; 66 : 589 – 600 .
43. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome . Cell . 2004 ; 119 : 847 – 860 .
44. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting . Science . 1994 ; 265 : 103 – 106 .
45. Engineering de novo reciprocal chromosomal translocations associated with Mll to replicate primary events of human cancer . Cancer Cell . 2003 ; 3 : 449 – 458 .
46. Mll fusions generated by Cre- loxP-mediated de novo translocations can induce lineage reassignment in tumorigenesis . EMBO J . 2005 ; 24 : 3136 – 3146 .
47. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras . Genes Dev . 2001 ; 15 : 3243 – 3248 .
48. Nf2 gene inactivation in arachnoidal cells is rate-limiting for meningioma development in the mouse . Genes Dev . 2002 ; 16 : 1060 – 1065 .
49. Role of K-ras and Pten in the development of mouse models of endometriosis and endometrioid ovarian cancer . Nat Med . 2005 ; 11 : 63 – 70 .
50. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model . Cancer Cell . 2003 ; 4 : 181 – 189 .
51. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase . Nat Protoc . 2009 ; 4 : 1064 – 1072 .
52. Surfing the p53 network . Nature . 2000 ; 408 : 307 – 310 .
53. Detection of mutant p53 in clam leukemia cells . Exp Cell Res . 1997 ; 232 : 240 – 245 .
54. Micro RNA expression profiles classify human cancers . Nature . 2005 ; 435 : 834 – 838 .
55. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene . Cell . 2006 ; 125 : 1269 – 1281 .
56. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach . Cell . 2006 ; 125 : 1253 – 1267 .
57. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis . Nat Genet . 2005 ; 37 : 48 – 55 .
58. Suppression of lung adenocarcinoma progression by Nkx2-1 . Nature . 2011 ; 473 : 101 – 104 .
59. The consensus coding sequences of human breast and colorectal cancers . Science . 2006 ; 314 : 268 – 274 .
60. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib . N Engl J Med . 2004 ; 350 : 2129 – 2139 .
61. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy . Science . 2004 ; 304 : 1497 – 1500 .
62. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor . J Clin Oncol . 2003 ; 21 : 4342 – 4349 .
63. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia . N Engl J Med . 2001 ; 344 : 1031 – 1037 .
64. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes . Nature . 1983 ; 304 : 596 – 602 .
65. EphB receptor activity suppresses colorectal cancer progression . Nature . 2005 ; 435 : 1126 – 1130 .
66. Genetic analysis of Pten andTsc2 functional interactions in the mouse reveals asymmetrical haploinsufficiency in tumor suppression . Genes Dev . 2005 ; 19 : 1779 – 1786 .
67. The deficiency of Aktl is sufficient to suppress tumor development in Pten mice . Genes Dev . 2006 ; 20 : 1569 – 1574 .
68. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma . Genes Dev . 2006 ; 20 : 2096 – 2109 .
69. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF . Genes Dev . 1999 ; 13 : 2678 – 2690 .
70. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer . Nat Genet . 2002 ; 32 : 153 – 159 .
71. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice . Cell . 2005 ; 122 : 473 – 483 .
72. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse . Nature . 2005 ; 436 : 272 – 276 .
73. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system . Nature . 2005 ; 436 : 221 – 226 .
74. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity . Cell . 2005 ; 121 : 849 – 858 .
75. A genetic screen for candidate tumor suppressors identifies REST . Cell . 2005 ; 121 : 837 – 848 .
76. A loss-of-function RNA interference screen for molecular targets in cancer . Nature . 2006 ; 441 : 106 – 110 .
77. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in E mu-myc transgenic mice . Cell . 1991 ; 65 : 753 – 763 .
78. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging . Cell . 1991 ; 65 : 737 – 752 .
79. New genes involved in cancer identified by retroviral tagging . Nat Genet . 2002 ; 32 : 166 – 174 .
80. Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model . Cancer Cell . 2009 ; 16 : 324 – 335 .
81. In vivo RNAi screening identifies regulators of actin dynamics as key determinants of lymphoma progression . Nat Genet . 2009 ; 41 : 1133 – 1137 .
82. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer . Cell . 2008 ; 135 : 852 – 864 .
83. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer . Science . 2009 ; 324 : 1457 – 1461 .
84. Assessing therapeutic responses in Kras mutant cancers using genetically engineered mouse models . Nat Biotechnol . 2010 ; 28 : 585 – 593 .
85. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response . Nature . 2012 ; 483 : 613 – 617 .
86. Lung cancer signatures in plasma based on proteome profiling of mouse tumor models . Cancer Cell . 2011 ; 20 : 289 – 299 .
87. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells . Cell . 2004 ; 119 : 431 – 443 .
88. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung . Cancer Cell . 2011 ; 19 : 754 – 764 .
89. Identification of bronchioalveolar stem cells in normal lung and lung cancer . Cell . 2005 ; 121 : 823 – 835 .
90. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis . Cell . 1996 ; 86 : 353 – 364 .
91. Stromal fibroblasts in cancer initiation and progression . Nature . 2004 ; 432 : 332 – 337 .
92. Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha . Science . 2010 ; 330 : 827 – 830 .
93. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene . Cell . 1988 ; 54 : 105 – 115 .
94. Stochastic appearance of mammary tumors in transgenic mice carrying the MMTV/c-neu oncogene . Cell . 1989 ; 57 : 931 – 936 .
95. Prostate cancer in a transgenic mouse . Proc Natl Acad Sci U S A . 1995 ; 92 : 3439 – 3443 .
96. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene . Science . 1992 ; 256 : 668 – 670 .
97. Loss of Apc heterozygosity and abnormal tissue building in nascent intestinal polyps in mice carrying a truncated Apc gene . Proc Natl Acad Sci U S A . 1995 ; 92 : 4482 – 4486 .
98. A targeted chain-termination mutation in the mouse Apc gene results in multiple intestinal tumors . Proc Natl Acad Sci U S A . 1994 ; 91 : 8969 – 8973 .
99. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice . Cancer Cell . 2005 ; 7 : 469 – 483 .
100. The differential effects of mutant p53 alleles on advanced murine lung cancer . Cancer Res . 2005 ; 65 : 10280 – 10288 .
101. Loss of transforming growth factor-beta type II receptor promotes metastatic head-and-neck squamous cell carcinoma . Genes Dev . 2006 ; 20 : 1331 – 1342 .