Chapter 31 Gene Array Technology and the Search for Cosmeceutical Actives
INTRODUCTION
1. The use of a rigorous cell and molecular biology-based screening program to identify active compounds with the desired biologic activity (e.g. collagen I, III, or VII gene stimulation).
2. The application of this screening program to determine that the identified ‘active’ ingredient does not also produce undesirable biologic effects on skin cells, e.g. stimulate matrix metalloproteinase 1 (MMP-1) gene activity.
3. The development of topical formulations which can be shown by skin percutaneous absorption analysis to deliver sufficient amounts of the ‘active’ ingredient across the stratum corneum and down to the target cells to achieve the required biologic effect.
4. The use of double-blind, placebo-controlled clinical studies with a sufficient number of patients to generate statistically significant data on product efficacy.
BASIC PRINCIPLES OF GENE ARRAY ANALYSIS
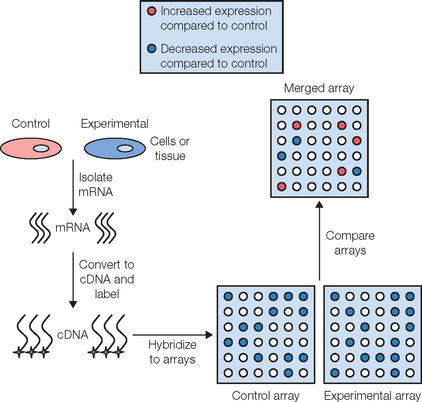
The sequence of steps involved in a gene array analysis is shown in Figure 31.1. The first step involves isolating mRNA from cells that represent the ‘control’ group, and from cells exposed to some experimental condition, such as UVR (‘experimental’ group). The mRNA preparation from each group is then reverse transcribed into ‘complementary DNA’ (cDNA), which is more stable and hybridizes better to DNA than mRNA. This cDNA is then labeled with either a radioisotope or a fluorescent tag so that each unique cDNA can be detected and identified at the conclusion of the experiment. Once the cDNAs have been tagged, they are incubated with the gene array filter (e.g. the ‘skin specific’ array) so that hybridization between a given cDNA and its complementary DNA on the array can occur. Once hybridization is complete, any unbound cDNA is washed away and the hybridized cDNA is detected and quantified. Since the location and identity of each gene on the filter is known, by comparing the quantified spots on the array produced from the ‘control’ group to those spots that are produced in the ‘experimental’ array, one can determine if a particular gene in the experimental group is upregulated or downregulated relative to the control group. Given the complexity of gene arrays, a computer software program is used to aid in the quantification and analysis of the large amount of data that is obtained. The software produces an ‘overlay’ image of both gene array filters, calculates the difference in expression level for each gene between the control and experimental groups, and then converts this relative expression data into a color image. Typically, a gene that is upregulated in the experimental group relative to the control group is shown as a green spot on the computer-generated image while genes that are downregulated are shown in red. An example of the use of this technology in the identification of a novel antiaging and anti-inflammatory active is discussed below.
APPLICATION OF GENE ARRAYS TO THE IDENTIFICATION AND CHARACTERIZATION OF ANTIAGING AND ANTI-INFLAMMATORY BIOACTIVE MOLECULES
As results from microarray technology have become more reliable and reproducible over the last few years, it has become possible to determine the effects of candidate ‘bioactive’ molecules on skin cells with increased confidence. Furthermore, the use of microarrays has expanded from basic research studies for candidate compound identification to screening tissue samples in a clinical setting to determine an individual’s susceptibility to certain diseases such as cancer. Due to the vast amount of data obtained from one particular experiment (e.g. 4000 genes of interest on one DNA filter), it is advantageous to only select a highly restricted set of ‘critical’ genes of interest for investigation. For example, if anti-inflammatory activity is desired one might investigate the regulation of genes such as COX-2 (PGE2 producing gene), IL-1α, IL-6, IL-8, and TNF-α. Alternatively, if an antiaging bioactive was desired one would focus on the expression of extracellular matrix genes such as collagens, elastin, and proteoglycans combined with the inhibition of matrix degrading proteases such as collagenase and the gelatinases. Microarray technology also offers the opportunity to identify new beneficial effects that would not have been discovered otherwise using typical single gene experiments.
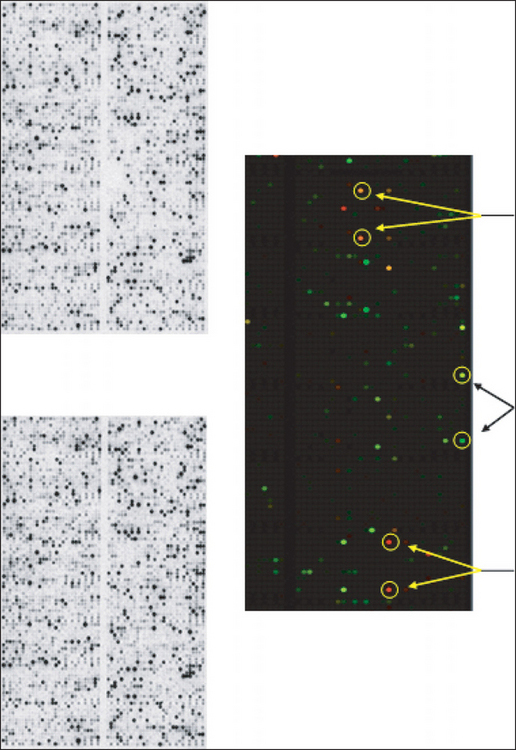
We have used gene array technology to identify unique compounds that have antiaging and/or anti-inflammatory effects in multiple skin cell types. In one study we assessed the ability of a novel nitrone spin-trap compound to modulate the expression of aging-related genes in older human dermal fibroblasts. The fibroblasts were grown in the presence (experimental) or absence (control) of the spin-trap nitrone compound, CX-412, for 48 hours at which time the mRNA from each cell culture group was isolated, converted to a complementary DNA (cDNA), labeled with radioactive nucleotides, and hybridized to IntegriDerm DermArray gene filters. These microarray filters contain over 4400 unique cDNAs specifically chosen due to their expression in skin cells and relevance to dermatologic research. Genes that are commonly expressed in skin are spotted in duplicate at different sites on the gene filters to provide an estimate of reproducibility of the hybridization reaction. The hybridization images are imported into a computer program that normalizes the data set and provides a color-coded picture of which expressed genes in the CX-412 treated fibroblasts were upregulated (coded in green) or downregulated (coded in red) relative to the untreated fibroblast cultures (Figure 31.2C). Figures 31.2A and B show the actual filter images of the hybridized radioactive cDNAs which were upregulated or downregulated by the spin-trap, CX-412.
After quantifying the level of all expressed genes in control and CX-412 treated cells, we found that aged fibroblasts treated with CX-412 shifted their expression patterns from matrix destruction to matrix production (Box 31.1). For example, inhibition of the matrix metalloproteinases collagenase 1 and 92 kDa gelatinase was noted, while the naturally occurring inhibitors of MMPs, TIMP-1 and TIMP-2, were upregulated. Furthermore, the expression of collagen types I, II, and III were enhanced in fibroblasts. In addition to age-related genes we also found that certain inflammation-associated genes, including uPA, tPA, PAI-1, IL-1α, and IL-6 were markedly inhibited by the spin-trap compound tested.
Blumenberg M. DNA microarrays in dermatology and skin biology. OMICS: a journal of integrative biology. 2006;10:243–260.
Cheepala SB, Syed Z, Trutschi M, Cvek U, Clifford JL. Retinoids and skin: microarrays shed new light on chemopreventive action of all-trans retinoic acid. Molecular Carcinogenesis. 2007;46:634–639.
Davis RLJr, DuBreuil RM, Reddy SP, Dooley TP. Methods for gene expression profiling in dermatology research using DermArray nylon filter DNA microarrays. Methods in Molecular Biology. 2005;289:399–412.
Floyd RA. Nitrones as therapeutics in age-related diseases. Aging Cell. 2006;5:51–57.
Fore J. A review of skin and the effects of aging on skin structure and function. Ostomy Wound Management. 2006;52:24–35.
Landau M. Exogenous factors in skin aging. Current Problems in Dermatology. 2007;35:1–13.
Ramos-e-Silva M, da Silva Carneiro SC. Elderly skin and its rejuvenation: products and procedures for the aging skin. Journal of Cosmetic Dermatology. 2007;6:40–50.
Sellheyer K, Belbin TJ. DNA microarrays: from structural genomics to functional genomics. The applications of gene chips in dermatology and dermatopathology. Journal of the American Academy of Dermatology. 2004;51:681–692.