Enterobacteriaceae
1. Describe the general characteristics of the Enterobacteriaceae, including oxygenation, microscopic Gram staining characteristics, and macroscopic appearance on blood and MacConkey agar.
2. Describe the chemical principle of the media used for the isolation and differentiation of Enterobacteriaceae, including xylose-lysine-deoxycholate agar (XLD), Salmonella-Shigella agar (SS), Hektoen enteric agar (HE), MacConkey agar (MAC), eosin methylene blue agar (EMB), cefsulodin-irgasan-novobiocin agar (CIN), Simmons citrate agar (CIT), gram-negative broth (GN), MacConkey agar with sorbitol (MAC-SOR), lysine iron agar (LIA), and triple sugar iron agar (TSI).
3. Describe the antigens used for serotyping in Enterobacteriaceae, including bacterial location, chemical structure, heat stability, and nomenclature.
4. List the members of the Enterobacteriaceae that are considered intestinal pathogens (rather than extraintestinal pathogens).
5. Compare and contrast infections with the various pathotypes of Escherichia coli (i.e., uropathogenic E. coli [UPEC], meningitis/sepsis–associated E. coli [MNEC], enterotoxigenic E. coli [ETEC], enteroinvasive E. coli [EIEC], enteroaggregative E. coli [EAEC], enteropathogenic E. coli [EPEC], and enterohemorrhagic E. coli [EHEC]), including the route of transmission, types of infection, and pathogenesis.
6. Explain the clinical significance of E. coli O157:H7 and the recommended diagnostic testing for confirmation of infection.
7. Outline the basic biochemical testing procedure to differentiate Enterobacteriaceae from other gram-negative rods.
8. Define ESBL and interpret an antibiotic profile as either positive, negative for ESBL, including corrections required before reporting results.
9. Define MDRTF and the antibiotic susceptibility recommendations associated with identification of an MDRTF isolate.
10. Define an extended spectrum cephalosporin resistance and explain the clinical significance and identification in the clinical laboratory.
11. Describe the modified Hodge test (MHT) procedure, including the chemical principle and clinical significance of the test with regard to carbapenemase resistance.
12. Differentiate Salmonella spp. and Shigella spp. based on biochemical testing.
13. Differentiate Yersinia spp. from the major pathogens among the Enterobacteriaceae.
14. Correlate signs and symptoms of infection with the results of laboratory diagnostic procedures for the identification of a clinical isolate in the Enterobacteriaceae family.
Epidemiology
Enterobacteriaceae inhabit a wide variety of niches, including the human gastrointestinal tract, the gastrointestinal tract of other animals, and various environmental sites. Some are agents of zoonoses, causing infections in animal populations (Table 20-1). Just as the reservoirs for these organisms vary, so do their modes of transmission to humans.
TABLE 20-1
Epidemiology of Clinically Relevant Enterobacteriaceae
| Organism | Habitat (Reservoir) | Mode of Transmission |
| Escherichia coli | Normal bowel flora of humans and other animals; may also inhabit female genital tract | Varies with the type of infection. For nongastrointestinal infections, organisms may be endogenous or spread person to person, especially in the hospital setting. For gastrointestinal infections, the transmission mode varies with the strain of E. coli (see Table 20-2); it may involve fecal-oral spread between humans in contaminated food or water or consumption of undercooked beef or unpasteurized milk from colonized cattle |
| Shigella spp. | Only found in humans at times of infection; not part of normal bowel flora | Person-to-person spread by fecal-oral route, especially in overcrowded areas, group settings (e.g., daycare) and areas with poor sanitary conditions |
| Salmonella serotype Typhi Salmonella serotypes Paratyphi A, B, C |
Only found in humans but not part of normal bowel flora | Person-to-person spread by fecal-oral route by ingestion of food or water contaminated with human excreta |
| Other Salmonella spp. | Widely disseminated in nature and associated with various animals | Ingestion of contaminated food products processed from animals, frequently of poultry or dairy origin. Direct person-to-person transmission by fecal-oral route can occur in health care settings when hand-washing guidelines are not followed |
| Edwardsiella tarda | Gastrointestinal tract of cold-blooded animals, such as reptiles | Uncertain; probably by ingestion of contaminated water or close contact with carrier animal |
| Yersinia pestis | Carried by urban and domestic rats and wild rodents, such as the ground squirrel, rock squirrel, and prairie dog | From rodents to humans by the bite of flea vectors or by ingestion of contaminated animal tissues; during human epidemics of pneumonic (i.e., respiratory) disease, the organism can be spread directly from human to human by inhalation of contaminated airborne droplets; rarely transmitted by handling or inhalation of infected animal tissues or fluids |
| Yersinia enterocolitica | Dogs, cats, rodents, rabbits, pigs, sheep, and cattle; not part of normal human microbiota | Consumption of incompletely cooked food products (especially pork), dairy products such as milk, and, less commonly, by ingestion of contaminated water or by contact with infected animals |
| Yersinia pseudotuberculosis | Rodents, rabbits, deer, and birds; not part of normal human microbiota | Ingestion of organism during contact with infected animal or from contaminated food or water |
| Citrobacter spp., Enterobacter spp., Klebsiella spp., Morganella spp., Proteus spp., Providencia spp., and Serratia spp. | Normal human gastrointestinal microbiota | Endogenous or person-to-person spread, especially in hospitalized patients |
Pathogenesis and Spectrum of Diseases
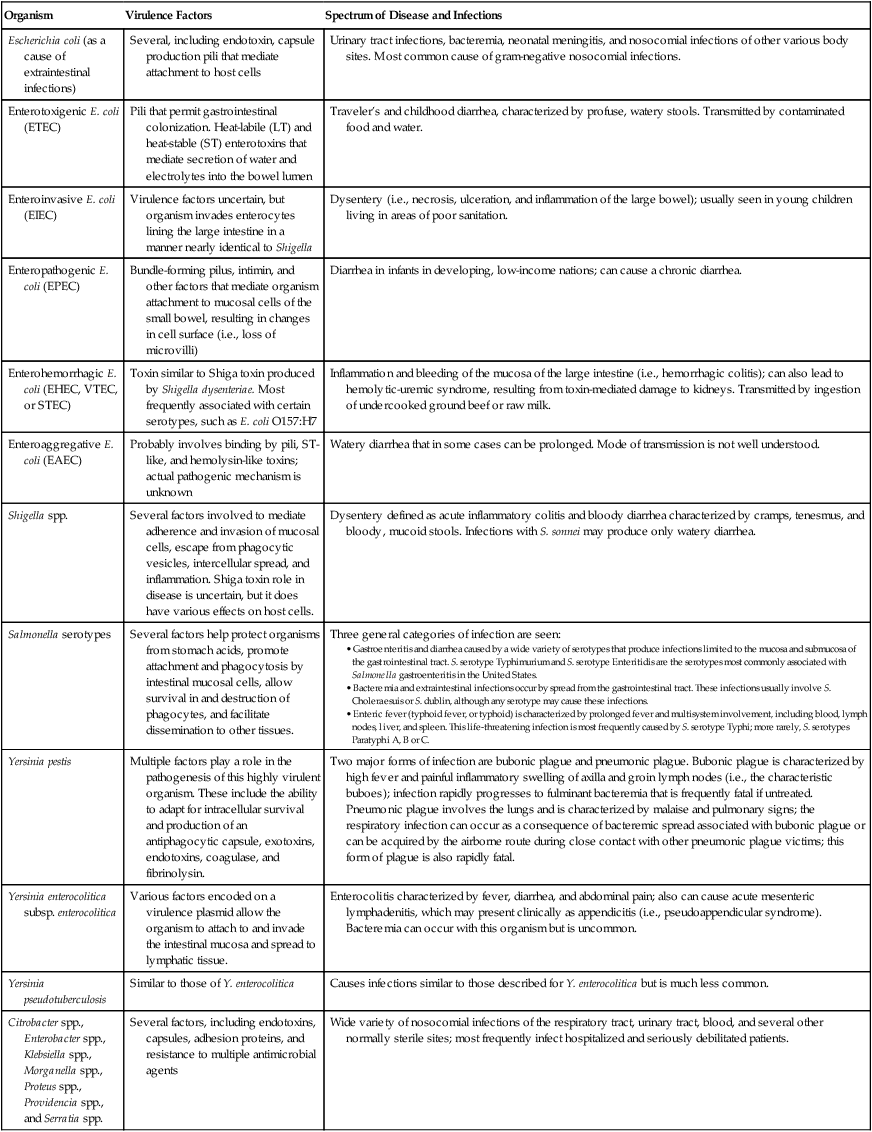
The clinically relevant members of the Enterobacteriaceae can be considered as two groups: the opportunistic pathogens and the intestinal pathogens. Typhi and Shigella spp. are among the latter group and are causative agents of typhoid fever and dysentery, respectively. Yersinia pestis is not an intestinal pathogen, but it is the causative agent of plague. The identification of these organisms in clinical material is serious and always significant. These organisms, in addition to others, produce various potent virulence factors and can cause life-threatening infections (Table 20-2).
TABLE 20-2
Pathogenesis and Spectrum of Disease for Clinically Relevant Enterobacteriaceae
| Organism | Virulence Factors | Spectrum of Disease and Infections |
| Escherichia coli (as a cause of extraintestinal infections) | Several, including endotoxin, capsule production pili that mediate attachment to host cells | Urinary tract infections, bacteremia, neonatal meningitis, and nosocomial infections of other various body sites. Most common cause of gram-negative nosocomial infections. |
| Enterotoxigenic E. coli (ETEC) | Pili that permit gastrointestinal colonization. Heat-labile (LT) and heat-stable (ST) enterotoxins that mediate secretion of water and electrolytes into the bowel lumen | Traveler’s and childhood diarrhea, characterized by profuse, watery stools. Transmitted by contaminated food and water. |
| Enteroinvasive E. coli (EIEC) | Virulence factors uncertain, but organism invades enterocytes lining the large intestine in a manner nearly identical to Shigella | Dysentery (i.e., necrosis, ulceration, and inflammation of the large bowel); usually seen in young children living in areas of poor sanitation. |
| Enteropathogenic E. coli (EPEC) | Bundle-forming pilus, intimin, and other factors that mediate organism attachment to mucosal cells of the small bowel, resulting in changes in cell surface (i.e., loss of microvilli) | Diarrhea in infants in developing, low-income nations; can cause a chronic diarrhea. |
| Enterohemorrhagic E. coli (EHEC, VTEC, or STEC) | Toxin similar to Shiga toxin produced by Shigella dysenteriae. Most frequently associated with certain serotypes, such as E. coli O157:H7 | Inflammation and bleeding of the mucosa of the large intestine (i.e., hemorrhagic colitis); can also lead to hemolytic-uremic syndrome, resulting from toxin-mediated damage to kidneys. Transmitted by ingestion of undercooked ground beef or raw milk. |
| Enteroaggregative E. coli (EAEC) | Probably involves binding by pili, ST-like, and hemolysin-like toxins; actual pathogenic mechanism is unknown | Watery diarrhea that in some cases can be prolonged. Mode of transmission is not well understood. |
| Shigella spp. | Several factors involved to mediate adherence and invasion of mucosal cells, escape from phagocytic vesicles, intercellular spread, and inflammation. Shiga toxin role in disease is uncertain, but it does have various effects on host cells. | Dysentery defined as acute inflammatory colitis and bloody diarrhea characterized by cramps, tenesmus, and bloody, mucoid stools. Infections with S. sonnei may produce only watery diarrhea. |
| Salmonella serotypes | Several factors help protect organisms from stomach acids, promote attachment and phagocytosis by intestinal mucosal cells, allow survival in and destruction of phagocytes, and facilitate dissemination to other tissues. | Three general categories of infection are seen:
• Gastroenteritis and diarrhea caused by a wide variety of serotypes that produce infections limited to the mucosa and submucosa of the gastrointestinal tract. S. serotype Typhimurium and S. serotype Enteritidis are the serotypes most commonly associated with Salmonella gastroenteritis in the United States. • Bacteremia and extraintestinal infections occur by spread from the gastrointestinal tract. These infections usually involve S. Choleraesuis or S. dublin, although any serotype may cause these infections. • Enteric fever (typhoid fever, or typhoid) is characterized by prolonged fever and multisystem involvement, including blood, lymph nodes, liver, and spleen. This life-threatening infection is most frequently caused by S. serotype Typhi; more rarely, S. serotypes Paratyphi A, B or C. |
Specific Organisms
Opportunistic Human Pathogens
Citrobacter spp. (C. freundii, C. koseri, C. braakii)
Citrobacter organisms are inhabitants of the intestinal tract. The most common clinical manifestation in patients as a result of infection occurs in the urinary tract. However, additional infections, including septicemias, meningitis, brain abscesses, and neurologic complications, have been associated with Citrobacter spp. Transmission is typically person to person. Table 20-3 provides an outline of the biochemical differentiation of the most common clinically isolated Citrobacter species. C. freundii may harbor inducible AmpC genes that encode resistance to ampicillin and first-generation cephalosporins.
TABLE 20-3
Biochemical Differentiation of Citrobacter Species
| Species | Indole | ODC | Malonate | ACID FERMENTATION | |||
| Adonitol | Dulcitol | Melibiose | Sucrose | ||||
| C. braakii | V | pos | neg | neg | V | V | neg |
| C. freundii | V | neg | neg | neg | neg | pos | V |
| C. koseri | pos | pos | pos | pos | V | neg | V |
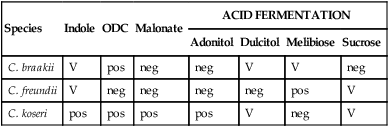
neg, Negative < 15%; ODC, ornithine decarboxylase; pos, positive ≥ 85%; V, variable 15% to 84%.
From Versalovic J: Manual of clinical microbiology, ed 10, 2011, Washington, DC, ASM Press.
Plesiomonas shigelloides
P. shigelloides is unusual in that it is among the few species of clinically relevant bacteria that decarboxylate lysine, ornithine, and arginine. It is important to distinguish Aeromonas spp. from P. shigelloides., since both are oxidase positive. This is accomplished by using the string test described in Chapter 26. The DNase test may also be used to differentiate these organisms. Aeromonas spp. are DNase positive and Plesiomonas organisms are DNase negative.
Laboratory Diagnosis
Specimen Collection and Transport
Enterobacteriaceae are typically isolated from a variety of sources in combination with other more fastidious organisms. No special considerations are required for specimen collection and transport of the organisms discussed in this chapter. (See Table 5-1 for general information on specimen collection and transport.)
Specimen Processing
No special considerations are required for processing of the great majority of organisms discussed in this chapter. The one exception is Yersinia pestis. This organism is a select agent. Manipulation of specimens suspected of containing this organism would generate aerosols and should be handled using Biosafety Level 3 (BSL-3) conditions. Refer to Table 5-1 for general information on specimen processing.
Cultivation
Media of Choice
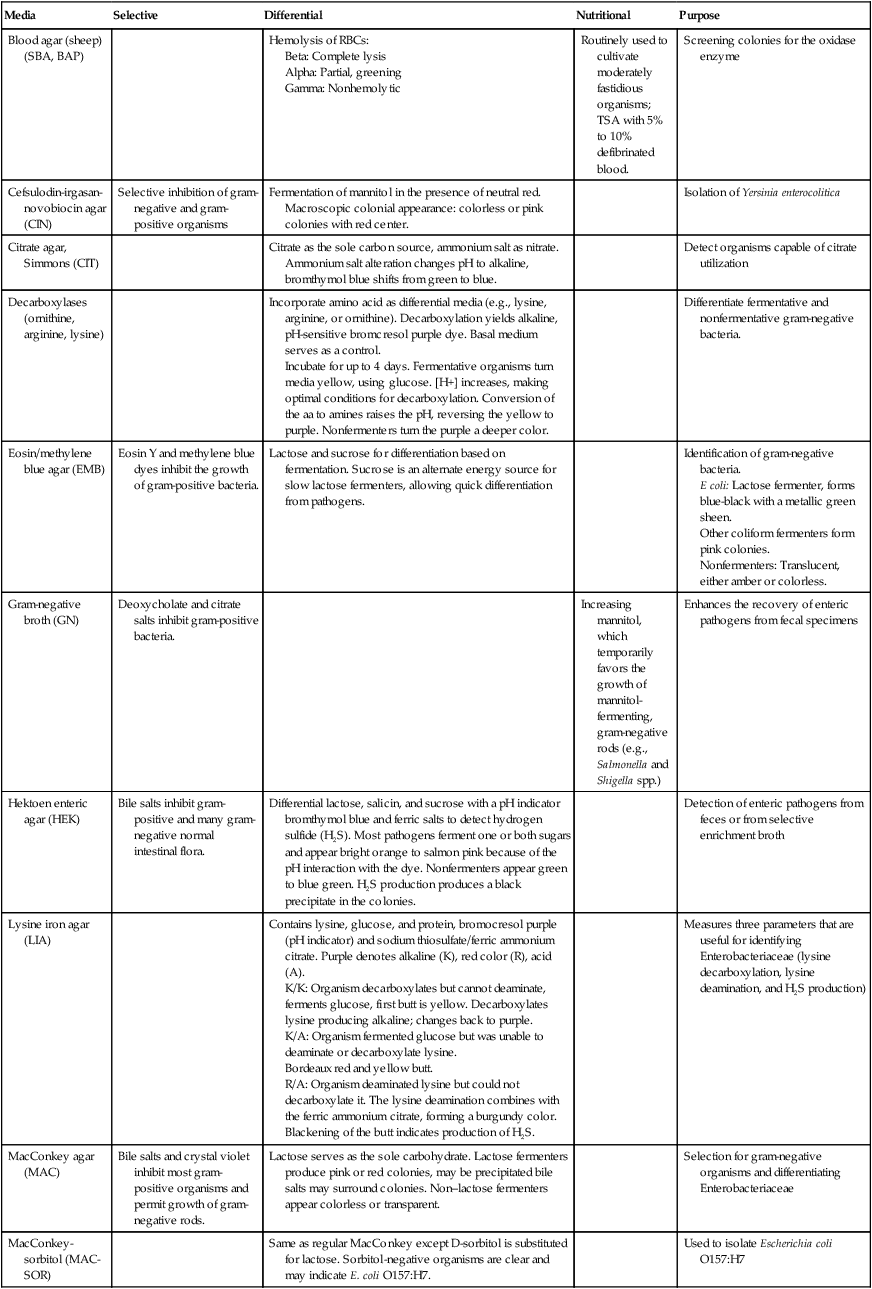
Most Enterobacteriaceae grow well on routine laboratory media, such as 5% sheep blood, chocolate, and MacConkey agars. In addition to these media, selective agars, such as Hektoen enteric (HE) agar, xylose-lysine-deoxycholate (XLD) agar, and Salmonella–Shigella (SS) agar, are commonly used to cultivate enteric pathogens from gastrointestinal specimens (see Chapter 59 for more information about laboratory procedures for the diagnosis of bacterial gastrointestinal infections). The broths used in blood culture systems, as well as thioglycollate and brain-heart infusion broths, all support the growth of Enterobacteriaceae.
Table 20-4 presents a complete description of the laboratory media used to isolate Enterobacteriaceae.
TABLE 20-4
Biochemical Media used in the Differentiation and Isolation of Enterobacteriaceae
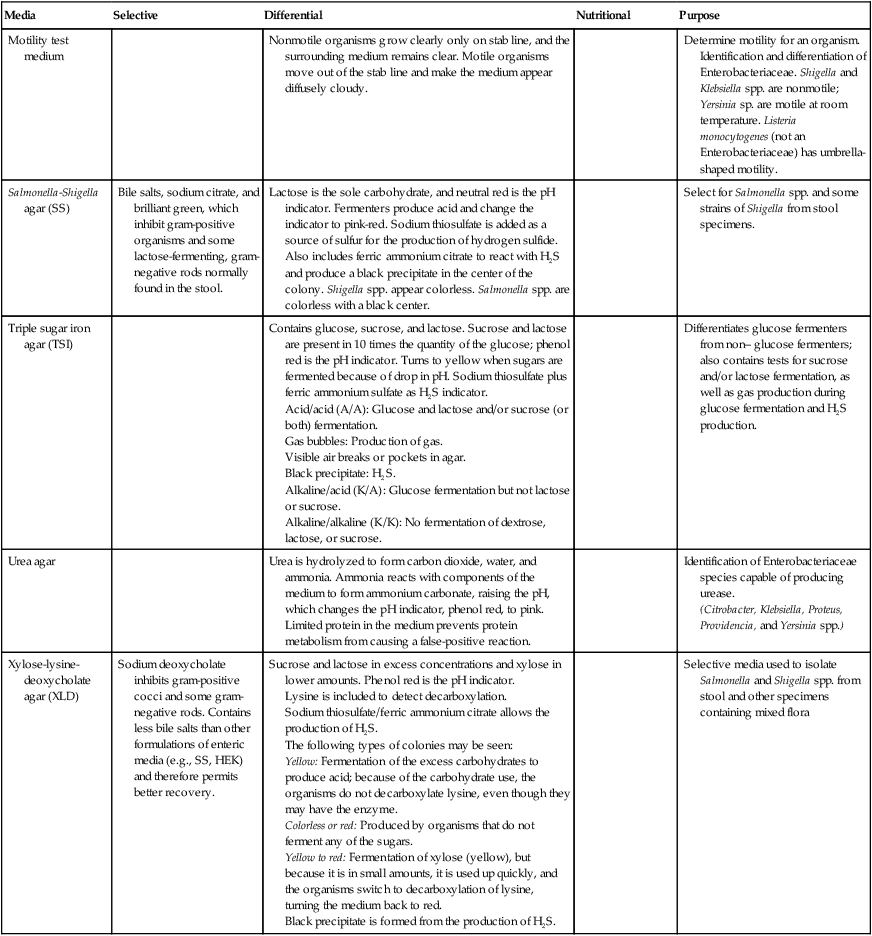
| Media | Selective | Differential | Nutritional | Purpose |
| Blood agar (sheep) (SBA, BAP) | Hemolysis of RBCs: | Routinely used to cultivate moderately fastidious organisms; TSA with 5% to 10% defibrinated blood. | Screening colonies for the oxidase enzyme | |
| Cefsulodin-irgasan-novobiocin agar (CIN) | Selective inhibition of gram-negative and gram-positive organisms | Fermentation of mannitol in the presence of neutral red. Macroscopic colonial appearance: colorless or pink colonies with red center. | Isolation of Yersinia enterocolitica | |
| Citrate agar, Simmons (CIT) | Citrate as the sole carbon source, ammonium salt as nitrate. Ammonium salt alteration changes pH to alkaline, bromthymol blue shifts from green to blue. | Detect organisms capable of citrate utilization | ||
| Decarboxylases (ornithine, arginine, lysine) | Incorporate amino acid as differential media (e.g., lysine, arginine, or ornithine). Decarboxylation yields alkaline, pH-sensitive bromcresol purple dye. Basal medium serves as a control. Incubate for up to 4 days. Fermentative organisms turn media yellow, using glucose. [H+] increases, making optimal conditions for decarboxylation. Conversion of the aa to amines raises the pH, reversing the yellow to purple. Nonfermenters turn the purple a deeper color. |
Differentiate fermentative and nonfermentative gram-negative bacteria. | ||
| Eosin/methylene blue agar (EMB) | Eosin Y and methylene blue dyes inhibit the growth of gram-positive bacteria. | Lactose and sucrose for differentiation based on fermentation. Sucrose is an alternate energy source for slow lactose fermenters, allowing quick differentiation from pathogens. | Identification of gram-negative bacteria. E coli: Lactose fermenter, forms blue-black with a metallic green sheen. Other coliform fermenters form pink colonies. Nonfermenters: Translucent, either amber or colorless. |
|
| Gram-negative broth (GN) | Deoxycholate and citrate salts inhibit gram-positive bacteria. | Increasing mannitol, which temporarily favors the growth of mannitol-fermenting, gram-negative rods (e.g., Salmonella and Shigella spp.) | Enhances the recovery of enteric pathogens from fecal specimens | |
| Hektoen enteric agar (HEK) | Bile salts inhibit gram-positive and many gram-negative normal intestinal flora. | Differential lactose, salicin, and sucrose with a pH indicator bromthymol blue and ferric salts to detect hydrogen sulfide (H2S). Most pathogens ferment one or both sugars and appear bright orange to salmon pink because of the pH interaction with the dye. Nonfermenters appear green to blue green. H2S production produces a black precipitate in the colonies. | Detection of enteric pathogens from feces or from selective enrichment broth | |
| Lysine iron agar (LIA) | Contains lysine, glucose, and protein, bromocresol purple (pH indicator) and sodium thiosulfate/ferric ammonium citrate. Purple denotes alkaline (K), red color (R), acid (A). K/K: Organism decarboxylates but cannot deaminate, ferments glucose, first butt is yellow. Decarboxylates lysine producing alkaline; changes back to purple. K/A: Organism fermented glucose but was unable to deaminate or decarboxylate lysine. Bordeaux red and yellow butt. R/A: Organism deaminated lysine but could not decarboxylate it. The lysine deamination combines with the ferric ammonium citrate, forming a burgundy color. Blackening of the butt indicates production of H2S. |
Measures three parameters that are useful for identifying Enterobacteriaceae (lysine decarboxylation, lysine deamination, and H2S production) | ||
| MacConkey agar (MAC) | Bile salts and crystal violet inhibit most gram-positive organisms and permit growth of gram-negative rods. | Lactose serves as the sole carbohydrate. Lactose fermenters produce pink or red colonies, may be precipitated bile salts may surround colonies. Non–lactose fermenters appear colorless or transparent. | Selection for gram-negative organisms and differentiating Enterobacteriaceae | |
| MacConkey-sorbitol (MAC-SOR) | Same as regular MacConkey except D-sorbitol is substituted for lactose. Sorbitol-negative organisms are clear and may indicate E. coli O157:H7. | Used to isolate Escherichia coli O157:H7 | ||
| Motility test medium | Nonmotile organisms grow clearly only on stab line, and the surrounding medium remains clear. Motile organisms move out of the stab line and make the medium appear diffusely cloudy. | Determine motility for an organism. Identification and differentiation of Enterobacteriaceae. Shigella and Klebsiella spp. are nonmotile; Yersinia sp. are motile at room temperature. Listeria monocytogenes (not an Enterobacteriaceae) has umbrella-shaped motility. | ||
| Salmonella–Shigella agar (SS) | Bile salts, sodium citrate, and brilliant green, which inhibit gram-positive organisms and some lactose-fermenting, gram-negative rods normally found in the stool. | Lactose is the sole carbohydrate, and neutral red is the pH indicator. Fermenters produce acid and change the indicator to pink-red. Sodium thiosulfate is added as a source of sulfur for the production of hydrogen sulfide. Also includes ferric ammonium citrate to react with H2S and produce a black precipitate in the center of the colony. Shigella spp. appear colorless. Salmonella spp. are colorless with a black center. | Select for Salmonella spp. and some strains of Shigella from stool specimens. | |
| Triple sugar iron agar (TSI) | Contains glucose, sucrose, and lactose. Sucrose and lactose are present in 10 times the quantity of the glucose; phenol red is the pH indicator. Turns to yellow when sugars are fermented because of drop in pH. Sodium thiosulfate plus ferric ammonium sulfate as H2S indicator. Acid/acid (A/A): Glucose and lactose and/or sucrose (or both) fermentation. Gas bubbles: Production of gas. Visible air breaks or pockets in agar. Black precipitate: H2S. Alkaline/acid (K/A): Glucose fermentation but not lactose or sucrose. Alkaline/alkaline (K/K): No fermentation of dextrose, lactose, or sucrose. |
Differentiates glucose fermenters from non– glucose fermenters; also contains tests for sucrose and/or lactose fermentation, as well as gas production during glucose fermentation and H2S production. | ||
| Urea agar | Urea is hydrolyzed to form carbon dioxide, water, and ammonia. Ammonia reacts with components of the medium to form ammonium carbonate, raising the pH, which changes the pH indicator, phenol red, to pink. Limited protein in the medium prevents protein metabolism from causing a false-positive reaction. | Identification of Enterobacteriaceae species capable of producing urease. (Citrobacter, Klebsiella, Proteus, Providencia, and Yersinia spp.) |
||
| Xylose-lysine-deoxycholate agar (XLD) | Sodium deoxycholate inhibits gram-positive cocci and some gram-negative rods. Contains less bile salts than other formulations of enteric media (e.g., SS, HEK) and therefore permits better recovery. | Sucrose and lactose in excess concentrations and xylose in lower amounts. Phenol red is the pH indicator. Lysine is included to detect decarboxylation. Sodium thiosulfate/ferric ammonium citrate allows the production of H2S. The following types of colonies may be seen: Yellow: Fermentation of the excess carbohydrates to produce acid; because of the carbohydrate use, the organisms do not decarboxylate lysine, even though they may have the enzyme. Colorless or red: Produced by organisms that do not ferment any of the sugars. Yellow to red: Fermentation of xylose (yellow), but because it is in small amounts, it is used up quickly, and the organisms switch to decarboxylation of lysine, turning the medium back to red. Black precipitate is formed from the production of H2S. |
Selective media used to isolate Salmonella and Shigella spp. from stool and other specimens containing mixed flora |
Incubation Conditions and Duration
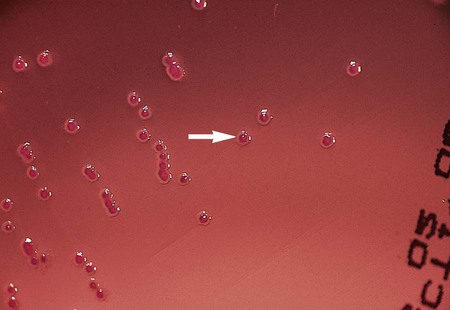
Under normal circumstances, most Enterobacteriaceae produce detectable growth in commonly used broth and agar media within 24 hours of inoculation. For isolation, 5% sheep blood and chocolate agars may be incubated at 35°C in carbon dioxide or ambient air. However, MacConkey agar and other selective agars (e.g., SS, HE, XLD) should be incubated only in ambient air. Unlike most other Enterobacteriaceae, Y. pestis grows best at 25° to 30°C. Colonies of Y. pestis are pinpoint at 24 hours but resemble those of other Enterobacteriaceae after 48 hours. CIN agar, used for the isolation of Y. enterocolitica, should be incubated 48 hours at room temperature to allow for the development of typical “bull’s-eye” colonies (Figure 20-1).
Colonial Appearance
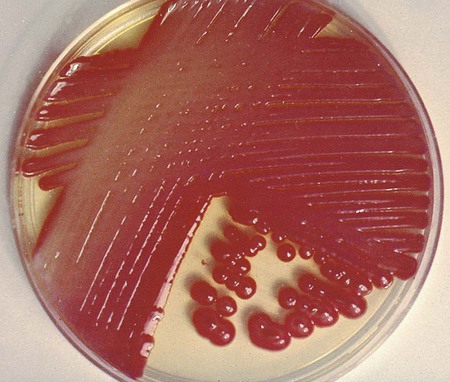
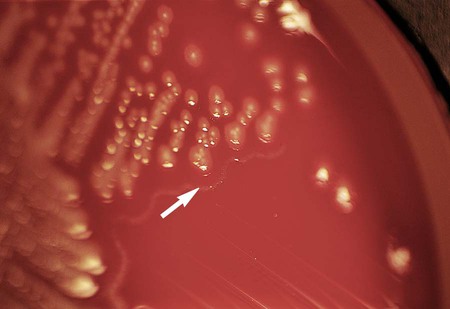
Table 20-5 presents the colonial appearance and other distinguishing characteristics (pigment and odor) of the most commonly isolated Enterobacteriaceae on MacConkey, HE, and XLD agars (see Figures 7-4, 7-6, and 7-8 for examples). All Enterobacteriaceae produce similar growth on blood and chocolate agars; colonies are large, gray, and smooth. Colonies of Klebsiella or Enterobacter may be mucoid because of their polysaccharide capsule. E. coli is often beta-hemolytic on blood agar, but most other genera are nonhemolytic. As a result of motility, Proteus mirabilis, P. penneri, and P. vulgaris “swarm” on blood and chocolate agars. Swarming results in the production of a thin film of growth on the agar surface (Figure 20-3) as the motile organisms spread from the original site of inoculation.
TABLE 20-5
Colonial Appearance and Characteristics of the Most Commonly Isolated Enterobacteriaceae*
| Organism | Medium | Appearance |
| Citrobacter spp. | MAC | Late LF; therefore, NLF after 24 hr; LF after 48 hr; colonies are light pink after 48 hr |
| HE | Colorless | |
| XLD | Red, yellow, or colorless colonies, with or without black centers (H2S) | |
| Edwardsiella spp. | MAC | NLF |
| HE | Colorless | |
| XLD | Red, yellow, or colorless colonies, with or without black centers (H2S) | |
| Enterobacter spp. | MAC | LF; may be mucoid |
| HE | Yellow | |
| XLD | Yellow | |
| Escherichia coli | MAC | Most LF, some NLF (some isolates may demonstrate slow or late fermentation); and generally flat, dry, pink colonies with a surrounding darker pink area of precipitated bile salts† |
| HE | Yellow | |
| XLD | Yellow | |
| Hafnia alvei | MAC | NLF |
| HE | Colorless | |
| XLD | Red or yellow | |
| Klebsiella spp. | MAC | LF; mucoid |
| HE | Yellow | |
| XLD | Yellow | |
| Morganella spp. | MAC | NLF |
| HE | Colorless | |
| XLD | Red or colorless | |
| Plesiomonas shigelloides | BAP | Shiny, opaque, smooth, nonhemolytic |
| MAC | Can be NLF or LF | |
| Proteus spp. | MAC | NLF; may swarm, depending on the amount of agar in the medium; characteristic foul smell |
| HE | Colorless | |
| XLD | Yellow or colorless, with or without black centers | |
| Providencia spp. | MAC | NLF |
| HE | Colorless | |
| XLD | Yellow or colorless | |
| Salmonella spp. | MAC | NLF |
| HE | Green, black center as a result of H2S production | |
| XLD | Red with black center | |
| Serratia spp. | MAC | Late LF; S. marcescens may be red pigmented, especially if plate is left at 25°C (Figure 20-2) |
| HE | Colorless | |
| XLD | Yellow or colorless | |
| Shigella spp. | MAC | NLF; S. sonnei produces flat colonies with jagged edges |
| HE | Green | |
| XLD | Colorless | |
| Yersinia spp. | MAC | NLF; may be colorless to peach |
| HE | Salmon | |
| XLD | Yellow or colorless |
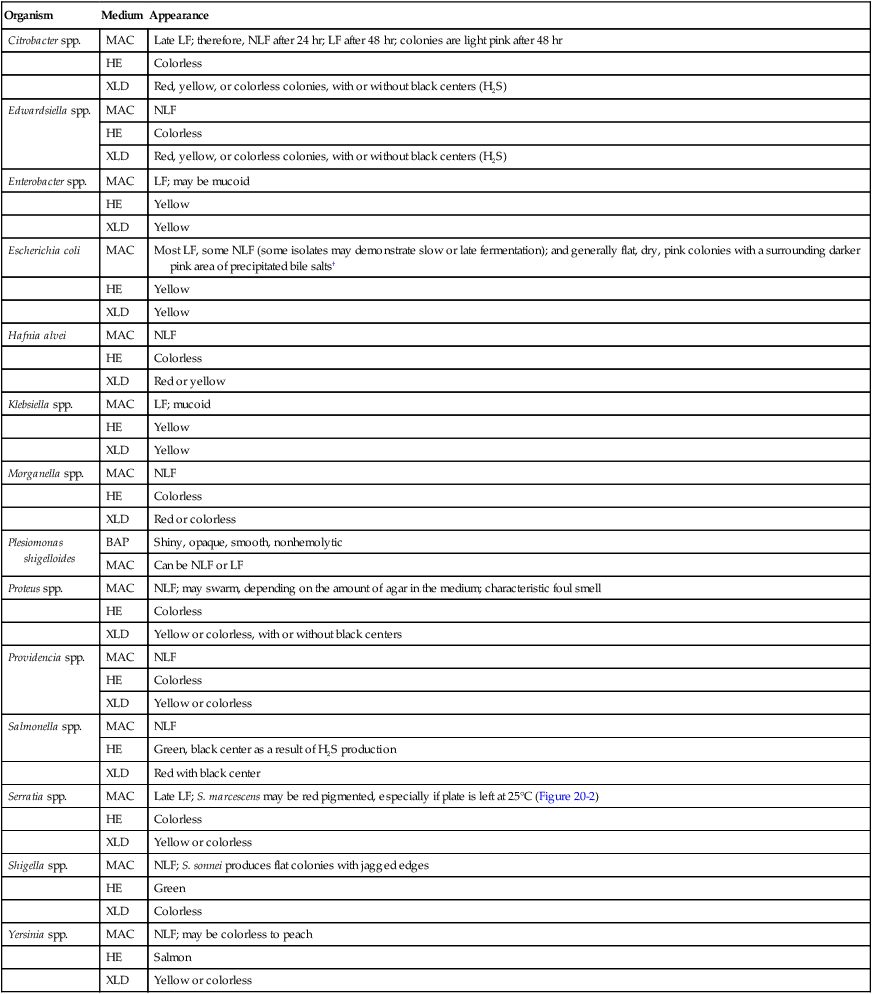
HE, Hektoen enteric agar; LF, lactose fermenter, pink colony; MAC, MacConkey agar; NLF, non–lactose fermenter, colorless colony; XLD, xylose-lysine-deoxycholate agar.
*Most Enterobacteriaceae are indistinguishable on blood agar; see text for colonial description.
†Pink colonies on MacConkey agar with sorbitol are sorbitol fermenters; colorless colonies are non–sorbitol fermenters.
Y. enterocolitica produces bull’s-eye colonies (dark red or burgundy centers surrounded by a translucent border; see Figure 20-1) on CIN agar at 48 hours. However, because most Aeromonas spp. produce similar colonies on CIN agar, it is important to perform an oxidase test to verify that the organisms are Yersinia spp. (oxidase negative). The oxidase test should be performed on suspect colonies that have been subcultured to sheep blood agar (Table 20-4). Pigments present in the CIN agar will interfere with correct interpretation of the oxidase test results.
Approach to Identification
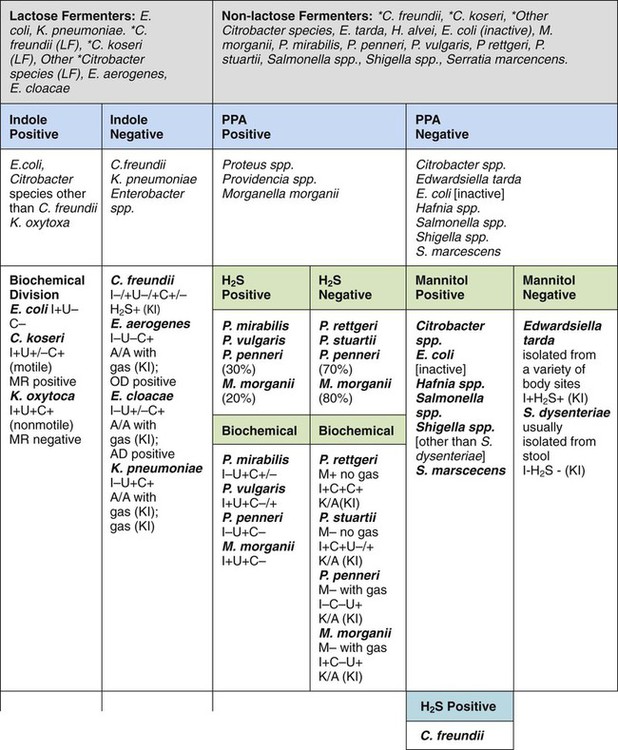
In the interests of cost containment, many clinical laboratories use an abbreviated scheme to identify commonly isolated enterics. E. coli, for example, the most commonly isolated enteric organism, may be identified by a positive spot indole test (see Procedure 13-41). For presumptive identification of an organism as E. coli, the characteristic colonial appearance on MacConkey agar, as described in Table 20-5, is documented along with positive spot indole test result. A spot indole test can also be used to quickly separate swarming Proteae, such as P. mirabilis and P. penneri, which are negative, from the indole-positive P. vulgaris.
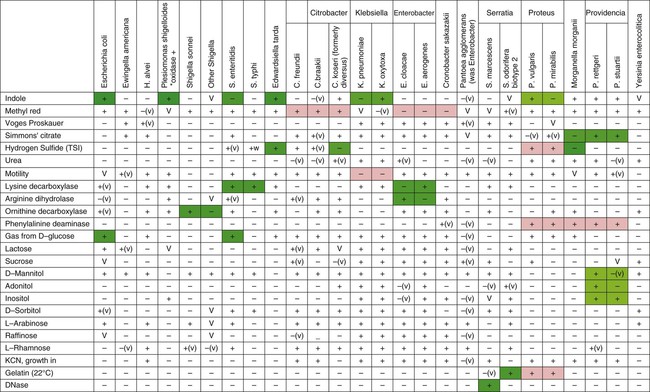
Table 20-6 provides an overview of common reactions for identifying biochemically unusual enteric pathogens. Figure 20-4 depicts the biochemical reactions typically used to differentiate some of the representative enteric pathogens. To aid the development of an understanding of the separation of common enteric pathogens based on groupings, Figure 20-5 provides a systematic algorithm for grouping pathogens into a working identification scheme.
TABLE 20-6
Biochemical Differentiation of Unusual LDC-, ODC- and ADH-negative Enterobacteriaceae
| Genus | Gas from Glucose | Motility | KCN | VP | ACID FERMENTATION | ||
| L-Arabitol | Sucrose | Trehalose | |||||
| Budvicia | neg | ||||||
| Ewingella | neg | V | neg | pos | neg | neg | pos |
| Leclercia | pos | pos | pos | neg | pos | pos | pos |
| Moellerella | pos | pos | V | neg | pos | neg | pos |
| Rahnella | pos | neg | neg | neg | neg | pos | pos |
| Tatumella | neg | neg | neg | neg | neg | pos | pos |
| Photorhabdus | neg | pos | neg | neg | neg | neg | neg |
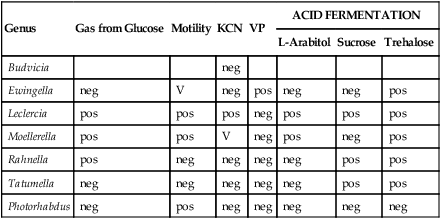
ADH, Arginine dihydrolase; KCN, potassium cyanide; LDC, lysine decarboxylase; ODC, ornithine decarboxylase; neg, negative 10%. pos, positive 90%; V, variable 11% to 89%; VP, Voges-Proskauer test.
Specific Considerations for Identifying Enteric Pathogens
The common biochemical tests used to differentiate the species in the genus Citrobacter are illustrated in Table 20-3.
Table 20-7 illustrates the use of biochemical profiles obtained with triple sugar iron (TSI) agar and lysine iron agar (LIA) to presumptively identify enteric pathogens (see Chapter 13 for information on the principles, performance, and interpretation of these tests). Organisms that exhibit the profiles shown in Table 20-7 require further biochemical profiling and, in the case of Salmonella spp. and Shigella spp., serotyping to establish a definitive identification. Bacterial species not considered capable of causing gastrointestinal infections give profiles other than those shown, but further testing may be required.
TABLE 20-7
TSI and LIA Reactions Used to Screen for Enteropathogenic Enterobacteriaceae and Aeromonas/Vibrio spp.*†
| TSI Reactions‡ | LIA Reactions‡ | Possible Identification |
K/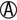 or K/A H2S + or K/A H2S + |
K/K or K/NC H2S+ | Salmonella serotypes Edwardsiella spp. |
| K/A H2S+ | K/K or K/NC H2S+ | Salmonella serotypes (rare) |
K/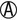 |
K/K or K/NC | Salmonella serotypes (rare) |
| K/A, H2S | K/K or K/NC H2S+ | Salmonella typhi (rare) |
K/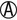 |
K/A H2S+ | Salmonella paratyphi A (usually H2S–) |
K/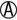 |
K/A or A/A | Escherichia coli Salmonella paratyphi A Shigella flexneri 6 (uncommon) Aeromonas spp. (oxidase positive) |
| K/A | K/K or K/NC | Plesiomonas sp. (oxidase positive) Salmonella typhi (rare) Vibrio spp. (oxidase positive) |
| K/A | K/A or A/A | Escherichia coli Shigella groups A-D Yersinia spp. |
A/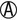 H2S+ H2S+ |
K/K or K/NC H2S+ | Salmonella serotypes (rare) |
| A/A | K/A or A/A | Escherichia coli Yersinia spp. Aeromonas spp. (oxidase positive) Vibrio cholerae (rare, oxidase positive) |
| A/A | K/K or K/NC | Vibrio spp. (oxidase positive) |
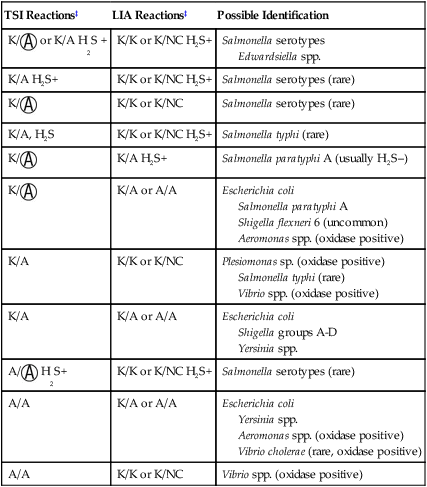
A, Acid; 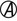 , acid and gas production; H2S, hydrogen sulfide; K, alkaline; LIA, lysine iron agar; NC, no change; TSI, triple sugar iron agar.
, acid and gas production; H2S, hydrogen sulfide; K, alkaline; LIA, lysine iron agar; NC, no change; TSI, triple sugar iron agar.
*Vibrio spp. and Aeromonas spp. are included in this table because they grow on the same media as the Enterobacteriaceae and may be enteric pathogens; identification of these organisms is discussed in Chapter 28.
†TSI and LIA reactions described in this table are only screening tests. The identity of possible enteric pathogens must be confirmed by specific biochemical and serologic testing.
‡Details regarding the TSI and LIA procedures can be found in Chapter 13.
P. shigelloides, a new member of the Enterobacteriaceae that can cause gastrointestinal infections (see Chapter 26), might cross-react with Shigella grouping antisera, particularly group D, and lead to misidentification. This mistake can be avoided by performing an oxidase test.
Antimicrobial Susceptibility Testing and Therapy
For many of the gastrointestinal infections caused by Enterobacteriaceae, inclusion of antimicrobial agents as part of the therapeutic strategy is controversial or at least uncertain (Table 20-8).
TABLE 20-8
Therapy for Gastrointestinal Infections Caused by Enterobacteriaceae
| Organisms | Therapeutic Strategies |
| Enterotoxigenic Escherichia coli (ETEC) Enteroinvasive E. coli (EIEC) Enteropathogenic E. coli (EPEC) Enterohemorrhagic E. coli (EHEC) Enteroaggregative E. coli (EAEC) | Supportive therapy, such as oral rehydration, is indicated in cases of severe diarrhea; for life-threatening infections, such as hemolytic-uremic syndrome associated with EHEC, transfusion and hemodialysis may be necessary. Antimicrobial therapy may shorten the duration of gastrointestinal illness, but many of these infections resolve without such therapy. Because these organisms may develop resistance (see Table 20-7), antimicrobial drug therapy for non–life-threatening infections may be contraindicated |
| Shigella spp. | Oral rehydration; antimicrobial drug therapy may be used to shorten the period of fecal excretion and perhaps limit the clinical course of the infection. However, because of the risk of resistance, using antimicrobial drug therapy for less serious infections may be questioned. |
| Salmonella serotypes | For enteric fevers (e.g., typhoid fever) and extraintestinal infections (e.g., bacteremia), antimicrobial agents play an important role in therapy. Potentially effective agents for typhoid include quinolones, chloramphenicol, trimethoprim/sulfamethoxazole, and advanced-generation cephalosporins, such as ceftriaxone; however, first- and second-generation cephalosporins and aminoglycosides are not effective. For nontyphoidal Salmonella bacteremia, a third-generation cephalosporin (e.g., ceftriaxone) is frequently used. For gastroenteritis, replacement of fluids is most important. Antimicrobial therapy generally is not recommended either for treatment of the clinical infection or for shortening the amount of time a patient excretes the organism. |
| Yersinia enterocolitica and Yersinia pseudotuberculosis | The need for antimicrobial therapy for enterocolitis and mesenteric lymphadenitis is not clear. In cases of bacteremia, pseudotuberculosis piperacillin, third-generation cephalosporins, aminoglycosides, and trimethoprim/sulfamethoxazole are potentially effective agents. Y. enterocolitica is frequently resistant to ampicillin and first-generation cephalosporins, whereas Y. pseudotuberculosis isolates are generally susceptible |
For extraintestinal infections, antimicrobial therapy is a vital component of patient management (Table 20-9). Although a broad spectrum of agents may be used for therapy against Enterobacteriaceae (see Table 12-6 for a detailed list), every clinically relevant species is capable of acquiring and using one or more of the resistance mechanisms discussed in Chapter 14. The unpredictable nature of any clinical isolate’s antimicrobial susceptibility requires that testing be done as a guide to therapy. As discussed in Chapter 12, several standard methods and commercial systems have been developed for this purpose. Table 20-10 presents intrinsic patterns of resistance identified in Enterobacteriaceae.
TABLE 20-9
Antimicrobial Therapy and Susceptibility Testing of Clinically Relevant Enterobacteriaceae
| Organism | Therapeutic Options | Potential Resistance to Therapeutic Options | Testing Methods* | Comments |
| Escherichia coli, Citrobacter spp., Enterobacter spp., Morganella spp., Proteus spp, Providencia spp., Serratia spp. | Several agents from each major class of antimicrobials, including aminoglycosides, beta-lactams, and quinolones, have activity. See Table 12-7 for a list of specific agents that should be selected for in vitro testing. For urinary tract infections, single agents may be used; for systemic infections, potent beta-lactams are used, frequently in combination with an aminoglycoside. | Yes; every species is capable of expressing resistance to one or more antimicrobials belonging to each drug class. | As documented in Chapter 12; disk diffusion agar dilution and commercial systems | In vitro susceptibility testing results are important for guiding broth dilution and therapy. |
| Yersinia pestis | Streptomycin is the therapy of choice; tetracycline and chloramphenicol are effective alternatives. | Yes, but rare | See CLSI document M100-515; testing must be performed only in a licensed reference laboratory. | Manipulation of cultures for susceptibility testing is dangerous for laboratory personnel and is not necessary. |
*Validated testing methods include standard methods recommended by the Clinical and Laboratory Standards Institute (CLSI) and commercial methods approved by the U.S. Food and Drug Administration (FDA).
TABLE 20-10
Intrinsic Antibiotic Resistance in Enterobacteriaceae*
| Escherichia hermannii | Hafnia alvei | Serratia marcescens | Yersinia enterocolitica | K. pneumoniae | CITROBACTER | ENTEROBACTER | PROTEUS | PROVIDENCIA | ||||||
| C. freundii | C. koseri | E. cloacae | E. aerogenes | P. vulgaris | P. mirabilis | P. penneri | P. rettgeri | P. stuartii | ||||||
| Ampicillin | R | R | R | R | R | R | R | R | R | R | R | R | R | |
| Amoxicillin/ clavulanate | R | R | R | R | R | R | R | R | R | |||||
| Ampicillin/sulbactam | R | R | R | R | R | R | ||||||||
| Piperacillin | R | |||||||||||||
| Ticarcillin | R | R | R | R | ||||||||||
| Cephalosporins I: cefazolin and cephalothin | R | R | R | R | R | R | R | R | R | R | ||||
| Cephamycins cefoxitin, and cefotetan | R | R | R | R | R | |||||||||
| Cephalosporins II: cefuroxime | R | R | R | R | R | R | ||||||||
| Tetracyclines | R | R | R | R | R | |||||||||
| Nitrofurantoin | R | R | R | R | R | R | ||||||||
| Polymyxin B Colistin |
R | R | R | R | R | R | ||||||||
*Cephalosporins III, cefepime, aztreonam, ticarcillin/clavulanate, piperacillin/tazobactam, and the carbapenems are not listed because the Enterobacteriaceae have no intrinsic resistance in to these antibiotics.
Modified from Clinical and Laboratory Standards Institute (CLSI): Performance standards for antimicrobial susceptibility testing—nineteenth informational supplement, CLS Document M100-S21, Wayne, Pa, 2011.
Extended Spectrum β-Lactamase (ESBL)–Producing Enterobacteriaceae
Enterobacteriaceae are capable of producing β-lactamases that hydrolyze penicillins and cephalosporins, including the extended spectrum cephalosporins (cefoxime, ceftriazone, ceftizoxime, and ceftazidime). These enzymes are referred to as ESBLs. A chromogenic agar has been developed for the detection of ESBLs. The agar chrom ID ESBL (bioMerieux, Marcy l’Etolle, France) uses cefpodoxime as a substrate to increase the recovery and sensitivity of CTX-M type ESBL isolates. Some limitations must be considered in the use of this medium, including hyperproducing AmpC (Enterobacter and Citrobacter spp.) and hyperproducing penicillinase (K. oxytoca) false positives. In addition, both Vitek 2 (bioMerieux, Durham, North Carolina) and Phoenix (Becton Dickinson, Sparks, Maryland) have ESBL panels, with expert interpretation available for clinical diagnostic use. Table 20-11 presents an example of an ESBL pattern from a clinical isolate that may require technical interpretation and correction before the results are reported.
TABLE 20-11
| Antibiotic | Vitek 2 | Expert* | Final† | |||
| Amikacin | 16 | S | 16 | S | 16 | S |
| Ampicillin | ≥32 | R | ≥32 | R | ≥32 | R |
| Ampicillin/sulbactam | ≥32 | R | ≥32 | R | ≥32 | R |
| Cefazolin | ≥64 | R | ≥64 | R | >-64 | R |
| Cefepime | 2 | S | 2 | R | 2 | R |
| Cefoxitin | ≥64 | R | ≥64 | R | ≥64 | R |
| Ceftazidime | ≥64 | R | ≥64 | R | ≥64 | R |
| Ceftriaxone | ≥64 | R | ≥64 | R | ≥64 | R |
| Ciprofloxacin | ≥4 | R | ≥4 | R | ≥4 | R |
| Ertapenem | ≤0.5 | S | ≤0.5 | S | ≤0.5 | S |
| ESBL | Pos | + | Pos | + | Pos | + |
| Gentamicin | ≤1 | S | ≤1 | S | ≤1 | S |
| Imipenem | ≤1 | S | ≤1 | S | ≤1 | S |
| Levofloxacin | ≥8 | R | ≥8 | R | ≥8 | R |
| Nitrofurantoin | ≤16 | S | ≤16 | S | ≤16 | S |
| Piperacillin/tazobactam | 16 | S | 16 | S | 16 | S |
| Tobramycin | ≥16 | R | ≥16 | R | ≥16 | R |
| Trimethoprim/sulfamethoxazole | ≤20 | S | ≤20 | S | ≤20 | S |
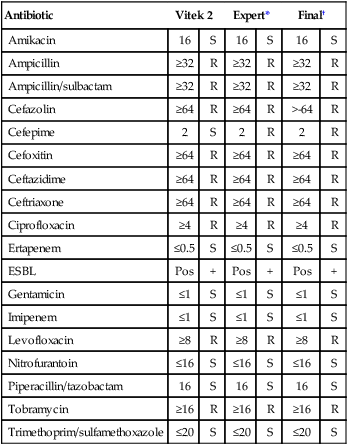
Suggested antibiogram correction: Therapeutic interpretations suggest corrections to cefepime; all other cephalosporins were already resistant.
Note: All the cephalosporins except cefepime display a resistance pattern.
*Expert findings indicate that susceptibility results are fully consistent with the organism identification.
†Final column indicates that the laboratory technologist corrected the interpretation as indicated before reporting the results to the physician.
Expanded-Spectrum Cephalosporin Resistance and carbapenemase resistance
The explosion of molecular biology in the past two decades has provided alternatives to phenotypic strategies for the identification of organisms and the genotyping of drug resistance. The bacterial chromosome represents the majority of the genetic make-up or genome within a single organism. However, many genes may be located on extra-chromosomal elements, including transposons and plasmids that are capable of independent replication and movement between organisms. Plasmids exist as double-stranded, closed, circular miniature chromosomes. A single bacterial cell may contain several plasmids. Transposable elements are pieces of DNA that move from one genetic element to another, such as from the plasmid to the chromosome or vice versa (see Chapter 2). Multi-drug resistant organisms are increasing in frequency on a worldwide basis due to the presence of these mobile genetic elements. In addition, these elements may have a complex structure, including the presence of integrans, which are genetic elements specifically designed to take up and incorporate or integrate genes such as those that encode antibiotic resistance.
1. While carbapenemase resistance may still be uncommon in some areas, at least 15 unusual biochemical resistance forms have been reported in the United States since July, 2012.
2. This increases the need for healthcare providers to work to aggressively prevent the emergence of CRE.
3. Guidelines are currently available from CDS to prevent CRE (e.g., contact precautions). Guidelines are available at http://www.cdc.gov/hai/organisms/cre/cre-toolkit/index.html.
4. Many of these organisms have been identified in patients within the United States following previous treatment and/or medication outside of the United States. These isolates should be referred to a reference laboratory for confirmatory susceptibility testing that should minimally include an evaluation for KPC and NDM carbapenemases.
1. All of the following organisms are considered normal intestinal floral except:
2. Enterobacteriaceae are typically gram negative and:
3. A patient presents with a urinary tract infection. After 24 hours of incubation, the urine culture grows a non–lactose fermenter on MacConkey agar, colorless colonies on HE indole-positive organism. The isolate is most likely:
4. Incubation of which organism at 25°C produces a characteristic yellow pigment?
5. The most common cause of hemolytic uremic syndrome is:
6. Which E. coli produces a heat-labile (LT) enterotoxin and a heat-stable enterotoxin?
7. A patient presents to the physician with pain and frequency of urination. The urine culture reveals a non–lactose fermenting, gram-negative rod with characteristic swarming on blood agar. The biochemical test that would specifically distinguish this organism from other Enterobacteriaceae is:
8. A patient presents with diarrhea and abdominal cramping. The organism isolated from the stool culture is identified as S. dysenteriae (group A). The TSI reaction would have indicated:
9. Which organism is commonly considered an extraintestinal pathogen?
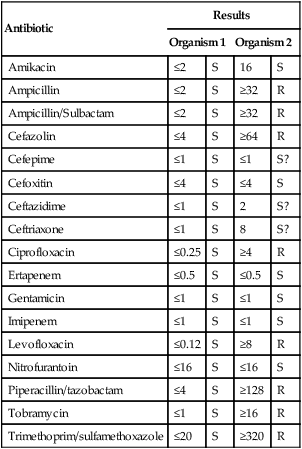
11. Short Answer: Interpret the following susceptibility patterns for two E. coli isolates.
| Antibiotic | Results | |||
| Organism 1 | Organism 2 | |||
| Amikacin | ≤2 | S | 16 | S |
| Ampicillin | ≤2 | S | ≥32 | R |
| Ampicillin/Sulbactam | ≤2 | S | ≥32 | R |
| Cefazolin | ≤4 | S | ≥64 | R |
| Cefepime | ≤1 | S | ≤1 | S? |
| Cefoxitin | ≤4 | S | ≤4 | S |
| Ceftazidime | ≤1 | S | 2 | S? |
| Ceftriaxone | ≤1 | S | 8 | S? |
| Ciprofloxacin | ≤0.25 | S | ≥4 | R |
| Ertapenem | ≤0.5 | S | ≤0.5 | S |
| Gentamicin | ≤1 | S | ≤1 | S |
| Imipenem | ≤1 | S | ≤1 | S |
| Levofloxacin | ≤0.12 | S | ≥8 | R |
| Nitrofurantoin | ≤16 | S | ≤16 | S |
| Piperacillin/tazobactam | ≤4 | S | ≥128 | R |
| Tobramycin | ≤1 | S | ≥16 | R |
| Trimethoprim/sulfamethoxazole | ≤20 | S | ≥320 | R |
Interpretation: _____________________________________________________________
___________________________________________________________________________
___________________________________________________________________________
___________________________________________________________________________
___________________________________________________________________________