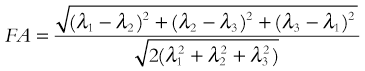Diffusion-Weighted Magnetic Resonance Imaging
Principles and Implementation in Clinical and Research Settings
Underlying Principles of DW-MRI Acquisition
Assuming that the movement of water molecules is not hindered by any form of barrier (so-called “free diffusion”), the mean squared distance that the spins will move over a given period is described by the Einstein-Smoluchowsky equation and is linearly proportional to the time (Δ) and to the self-diffusion coefficient D. The amount of attenuation is a function of the gradient strength G, gradient duration δ, time between gradient pulses Δ, and diffusion coefficient D. Typically G, δ, and Δ are combined to derive the “b-value”; the higher the b-value, the greater the signal attenuation in the resultant DW images (Fig. 26-1). In fact, signal attenuation is exponential, where S is the measured signal and S0 is the signal in the absence of diffusion weighting. As a result, D (measured in mm2/s) can be estimated by obtaining DW images at different b-values or by obtaining images with and without diffusion weighting.
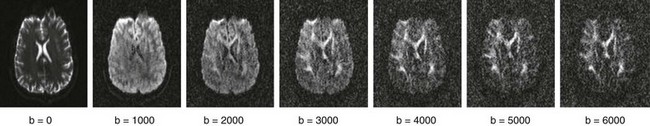
Figure 26-1 Changing b-value.
Displayed are corresponding slices from a single subject imaged at multiple b-values. Notice how contrast between gray and white matter increases with the increasing b-value, whereas overall signal/noise ratio decreases. (Data courtesy Justin Haldar, University of Southern California, Department of Electrical Engineering.)
Diffusion Weighting, Apparent Diffusivity, and Their Application in Clinical Settings
Clinically relevant information is available even from a single DWI acquisition. For instance, diffusion is restricted in regions of cytotoxic edema after a stroke, and these regions therefore will be hyperintense on DW images. However, typical DWI acquisitions are strongly T2-weighted, because a long echo time is necessary as a result of the time needed to apply the diffusion-sensitizing gradients. Therefore it is important to distinguish hyperintensity on DWI that represents true diffusion restriction from hyperintensity that reflects tissue T2 prolongation (often termed “T2 shine-through”). This differentiation usually is performed by the additional acquisition of an image without diffusion weighting (e.g., b = 0) and quantifying ADC on a pixel-by-pixel basis, which subsequently can be represented in gray scale as an ADC map. Additionally, instead of a single DW acquisition, three DW images typically are acquired using three orthogonal gradient directions, and the results are averaged to obtain directionally averaged DW images and ADC maps to minimize the effects of anisotropy. The directionally averaged ADC map is proportional to the trace of the diffusion tensor (described later), and thus the DWI images and ADC maps often are referred to as “trace-weighted DWI” and “trace diffusion tensor maps,” respectively (Fig. 26-2).
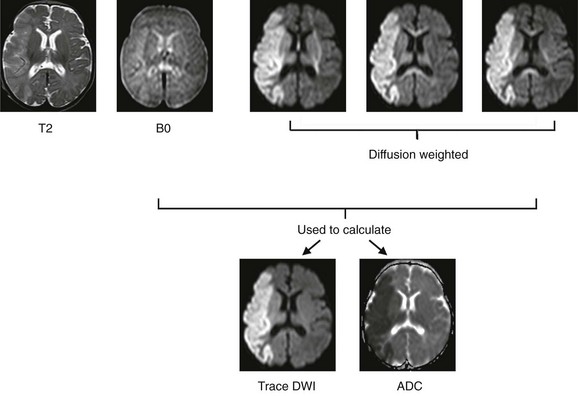
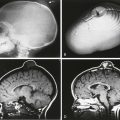
Figure 26-2 Diffusion-weighted (DW) imaging demonstrated in a 3-week-old neonate with an acute stroke.
Top row, Corresponding slices from an axial T2-weighted and DW-magnetic resonance imaging scan. In most protocols, three diffusion-encoding directions (rather than one) are acquired, averaged, and compared with the image without diffusion encoding (B0) to generate (bottom row) trace-weighted and apparent diffusivity maps that demonstrate areas of restricted diffusion as areas of high and low signal, respectively. Note that areas of directional anisotropy are visualized in each of the diffusion-encoding directions, but as a result of averaging, these areas are not apparent in the trace-weighted and apparent diffusion coefficient (ADC) maps.
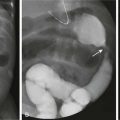
In addition to fluid homeostasis and intracellular water, DWI and ADC also are sensitive to the relative properties of water in the extracellular space. Thus DWI also is useful in the differential diagnosis of intracranial lesions. In lesions with high cellularity (e.g., high-grade tumors), the increased cellularity restricts water motion in the extracellular space, resulting in decreased ADC in the lesion (or in areas of the lesion with relatively higher cellularity). Accordingly, DWI often can distinguish high-grade tumors such as primitive neuroectodermal tumors from other lower grade pediatric brain tumors such as ependymomas or astrocytomas (Fig. 26-3).
Diffusion Tensor Imaging
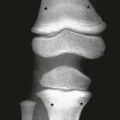
Although three diffusion-sensitizing gradients are sufficient for calculating a directionally averaged ADC image, a minimum of six directions is needed to characterize diffusion anisotropy. Anisotropic diffusion is directionally dependent, as within white matter fiber bundles, and is distinguishable from isotropic diffusion, which is observed in free fluids (e.g., the lateral ventricles) (Fig. 26-4).
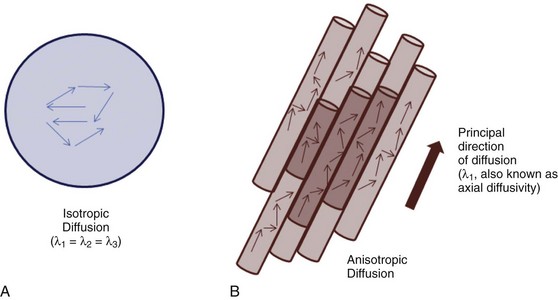
Figure 26-4 Isotropic and anisotropic diffusion.
A, Isotropic diffusion is exemplified by free diffusion such as in a large glass of water. B, Anisotropic diffusion occurs when diffusion is greater in one direction compared with the others, such as in axons. By convention, the principal eigenvalue (λ1) represents diffusion along the preferred axis.
As can be determined from this equation, FA can range from 0 (when λ1 = λ2 = λ3, i.e., isotropic diffusion) to 1 (when λ2 = λ3 = 0, i.e., fully anisotropic). In adults, typical white-matter values range between 0.9 and 0.4 depending on whether the measurement is obtained in a region where fiber bundles are heavily myelinated, tightly packed, and uniformly oriented (e.g., in the corpus callosum) or in a region where the organization deviates from that (such as in the vicinity of crossing fibers), respectively. FA values are much lower in neonates (ranging between 0.45 and 0.1) but rapidly increase toward the adult range in the first 2 years of life and then continue to increase at a much lower rate through adolescence into adulthood.1–3
To further enhance the clinical utility of FA, FA often is combined with information regarding the direction of the principal eigenvector, yielding a color FA map (Fig. 26-5). The typical convention is to color pixels red when the principal eigenvector is in the left-right direction relative to the 3D coordinate space, green when the principal eigenvector is anterior-posterior, and blue when the principal eigenvector is inferior-superior.
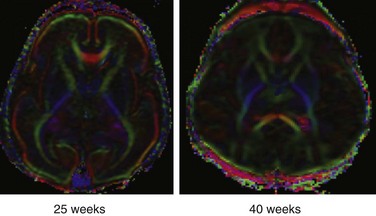
Figure 26-5 Color fractional anisotropy maps reconstructed from diffusion tensor imaging data acquired from a preterm neonate at 25 weeks and a term neonate at 40 weeks.
By convention, voxels in which the principal eigenvector (λ1) is in a left-right orientation (relative to the slice) are colored in red, voxels in which λ1 is in an anterior-posterior orientation are colored green, and voxels in which λ1 is in a superior-inferior orientation are colored blue. Note the presence of high anisotropy in the cortex of the neonate at 25 weeks and that, by term, the cortex has lower anisotropy than the adjacent white matter.
Microstructural Integrity
However, the precise interpretation for many of the differences in DTI measures of microstructure remains to be determined. Pioneering laboratory work by Sheng-Kwei Song and colleagues in mouse models demonstrated that myelin loss alone (without loss or degeneration of axons) results in an increase in radial diffusivity (RD), leaving axial diffusivity (AD) unchanged.4 In contrast, direct axonal damage results in a decrease in AD.5 Based on these findings, many researchers have drawn inverse conclusions on their own data—namely, that an increase in RD in a given white matter region reflects primarily damage to myelin, whereas a decrease in AD reflects primarily damage to axons. Unfortunately, the inverse conclusion is not necessarily valid. First, most of the laboratory work supporting the interpretation of AD and RD in relation to axonal pathology versus myelin has been carried out in the optic nerve, spinal cord, or corpus callosum.4–6 These structures are unique in the central nervous system in that they contain fiber bundles essentially aligned along a single axis. In contrast, the cerebrum and even the brainstem have far fewer regions where the fiber bundles are organized along a single axis (it has been estimated that as many as 90% of white matter voxels in the cerebrum contain crossing fibers).7 It is not known how altering myelination or axonal integrity along a single fiber bundle, in the setting of multiple crossing fibers, would alter AD or RD. Accordingly, in the cerebrum, given available knowledge at this point, a more appropriate interpretation may be “altered microstructure,” with more specific conclusions being drawn from collateral information. However, this technique continues to show great promise.
Tractography
Some of the increasingly common—and most important—questions that researchers and neuroradiologists have aimed to address with use of DTI concerns anatomic connectivity (e.g., “Which cortical and subcortical regions are connected, and by which fiber pathways?” “How strong are the connections between X and Y? … in this population compared with that one?”). To address these types of questions, most researchers and clinicians begin by constructing a visual representation of the white matter fibers (or tracts). This task is carried out with use of computer algorithms that regard fiber tracts as a continuous trajectory derived from local (voxel or pixel) estimates of fiber orientation (e.g., eigenvectors; Fig. 26-6). This technique is commonly known as tractography. It should be emphasized that no tractography method is capable of reconstructing axonal fibers or even fiber bundles. Rather, these methods compute trajectories or pathways represented by the data, which, it is hoped, parallel the predominant trajectories of the underlying axonal fiber bundles.
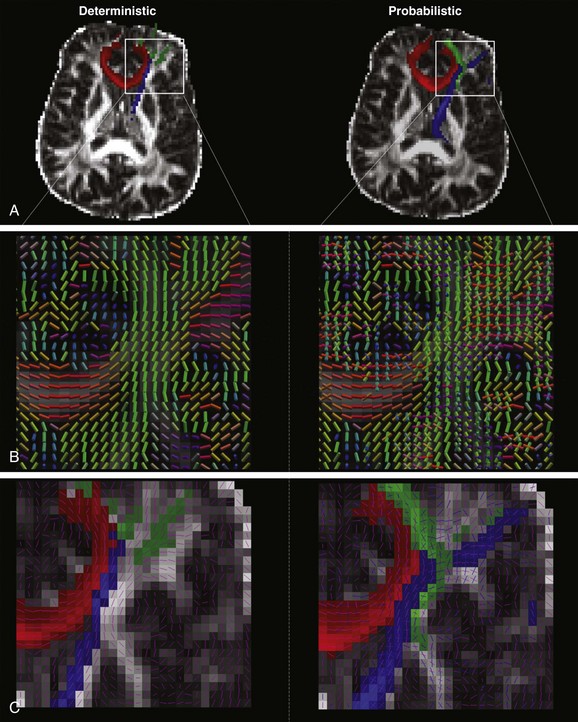
Figure 26-6 Deterministic and probabilistic tractography.
A, The same seed and waypoints were used to track anterior thalamic radiation (blue), genu of corpus callosum (red), and inferior frontooccipital fasciculus using deterministic (left) and probabilistic (right) algorithms on the same dataset acquired on a single subject. B, In deterministic tractography, the direction of anisotropy is considered to be along the axis of a single, principal eigenvector, which is represented voxelwise in the image on the left. (Color convention is as described in Fig. 26-5.) In contrast, probabilistic tractography models anisotropy as a probability distribution and allows for a two-fiber solution, which is represented on the right, with the major axis being larger in scale compared with the minor axis. C, The difference in output is clearly visualized in the region of the crossing fibers where, on the left, deterministic tractography (principal eigenvector overlaid as pink lines) fails to yield as many tracks as the output from the probabilistic algorithm on the right (again, principal and secondary eigenvectors are overlaid in pink and blue, respectively).
One of the most commonly used algorithms for tractography is known as fiber assignment by continuous tracking (also known as deterministic tractography).8 This algorithm proceeds from an initially determined point (seed region) and propagates pixel by pixel in the direction of the principal eigenvector until a predetermined lower FA threshold or maximal turning angle is reached, at which point the fiber path is terminated (see Fig. 26-6). The result is a streamline, a visual representation of anisotropic diffusion, and importantly, not an actual visual representation of axons or fiber bundles. Moreover, the number of streamlines originating from a particular seed region is directly proportional to the number of seed points within a given voxel, and accordingly, not directly related to the underlying anatomic connectivity. However, assuming the same number of seed points per given voxel are used across populations of interest (which, in most software packages, is not a number that can be manipulated by the operator), the number of streamlines can be considered a proxy for the underlying tissue microstructure, and accordingly, may be used as a metric in group-level comparisons.
One of the key limitations in tractography is that the mathematical model used to reconstruct DTI data assumes that a single fiber population exists in each voxel. However, at the resolution of a typical DTI acquisition (≥2 mm in each dimension), many voxels contain populations of fibers that are oriented in some manner other than parallel (from one third to 90%),7,8 including fibers that are crossing, fanning, branching, or bottlenecking. This crossing fiber problem is a significant concern if one is trying to carry out tractography on DTI data. First, the main effect of crossing or other nonparallel fiber orientations on the principal eigenvectors is to decrease the principal eigenvalue. This effect, in turn, results in a decrease in FA, which can cause tractography algorithms to prematurely stop if the FA value has fallen below the predetermined threshold. Second, because the algorithm follows only the principal eigenvector, it may generate spurious streamlines (e.g., a streamline that propagates into a crossing region perpendicular to the principal eigenvector could be made to bend and then continue in the new direction).
One method to address limitations associated with using deterministic tractography and DTI parameters is to use the information derived from the diffusion tensor fit to generate a 3D probability distribution regarding the diffusion of water molecules instead of constraining the molecules to only move along the direction of the principal eigenvector. In this way, probabilistic tractography algorithms allow water molecules to follow more than one orientation when passing through a voxel in a crossing region. Current probabilistic algorithms allow for the modeling of uncertainty of two (or more) fiber directions at each voxel. Tracking is done by launching a high number of streamlines from a seed region, and at each voxel—instead of deterministically following the principal eigenvector—drawing from the previously determined 3D probability distribution to propagate the tracts. Coherent fiber orientation is preserved by following the sampled direction that is most closely parallel to the previous location’s direction, as opposed to only following an arbitrary angle threshold. After a sufficient number of samples, the output is a probabilistic mapping of the uncertainty of fiber tracts at each voxel, with the dominant streamline surfacing as most probable (see Fig. 26-6). Notably, this method does not resolve the crossing-fiber problem with 100% certainty, but the resulting probabilistic map yields a much more robust estimate of the dominant fiber path when compared with deterministic approaches.9 Moreover, it also shows improvements in the rendering of the lateral corticospinal and corticobulbar tracts and the medial portion of the superior longitudinal fasciculus, which typically are not visualized by deterministic tractography. Figure 26-6 shows the comparison between deterministic and probabilistic algorithms in modeling fibers passing through a densely packed, high-fiber–crossing region of the brain. The probabilistic output shows significant improvement in delineating crossing fiber tracts, such as the inferior frontooccipital fasciculus and anterior thalamic radiation. Similar performance is noted in a single direction tract, as in the genu of the corpus callosum. Despite improved performance in modeling regions with crossing fiber tracts, probabilistic tractography still cannot completely overcome the limitation that the diffusion tensor model is not an adequate model for the underlying physiology in crossing fiber regions. This limitation requires more advanced approaches, as described in the next section.
Advanced Diffusion Imaging Techniques
To address the problem of crossing fibers, HARDI methods (sometimes called “Q-ball”)10,11 sample a much larger number of diffusion directions (e.g., 64 to 256) and then typical DTI acquisitions (e.g., ~30, although DTI analysis is possible with as few as 6). Thus more complicated models than a simple tensor may be used to model the physiology (for further detail, the interested reader is referred to references 11 to 14 for an introduction).12–15 It is possible, with these techniques, to model two, three, or even more crossing fiber bundles. In conjunction with probabilistic tractography, this provides a very powerful technique to model brain structural connectivity, and success has been seen even in regions with a very high number of crossing fibers. Typically, higher b-values are used in HARDI acquisitions (~3000 s/mm2) as opposed to those used in DTI (~1000 s/mm2). The reason is that diffusion is more restricted in directions perpendicular to the axon for intraaxonal water molecules, as opposed to interstitial water molecules, because the distance to the axonal membrane is smaller. Therefore the use of a higher b-value will suppress the signal from interstitial water, leading to a “cleaner” profile of fiber directionality.
Because most voxels consist of a mix of interstitial and intraaxonal water, the signal attenuation will deviate from an exponential decay with respect to b-value at higher b-values as the relative contribution from each type of water molecule changes. One approach to quantifying this deviation is DKI, which involves acquisition of data at several b-values; the “kurtosis” quantities describe deviations from exponential decay. (For details about DKI acquisition and analysis, see reference 16.) Compared with DTI, DKI has been shown to be more sensitive to subtle tissue microstructural changes17 and also has been found to yield additional information regarding pathologic changes in neural tissue, such as glioma grade discrimination.18 In typical DKI acquisitions, however, the angular resolution is not as great as HARDI because acquiring data at additional b-values is performed in lieu of additional diffusion gradient directions.
Finally, the “ultimate” in DWI acquisition is when acquisitions are obtained both at multiple gradient directions and multiple b-values,19 which will make possible a completely accurate specification of the water diffusion profiles, enabling the simultaneous modeling of crossing fiber bundles as well as nonexponential decay. This technique is called DSI or Q-space imaging. The acquisition demands are intense; a typical DSI protocol involves 512 separate DWI acquisitions, making the total acquisition time longer than 1 hour for typical DWI acquisition protocols.
Jones, DK, Knösche, TR, Turner, R. White matter integrity, fiber count and other fallacies: the do’s and don’ts of diffusion MRI. Neuroimage. 2012;pii:S1053–S8119. (12)01025-7, doi: 10.1016/j.neuroimage.2012.06.081 [E-pub ahead of print]
Roberts, TPL, Schwartz, ES. Principles and implementation of diffusion-weighted and diffusion tensor imaging. Pediatr Radiol. 2007;37:739–748.
References
1. Schmithorst, VJ, Wilke, M, Dardzinski, BJ, et al. Correlation of white matter diffusivity and anisotropy with age during childhood and adolescence: a cross-sectional diffusion-tensor MR imaging study. Radiology. 2002;222:212–218.
2. Schneider, JFL, Il’yasov, KA, Hennig, J, et al. Fast quantitative diffusion-tensor imaging of cerebral white matter from the neonatal period to adolescence. Neuroradiology. 2004;46:258–266.
3. Gao, W, Lin, G, Chen, Y, et al. Temporal and spatial development of axonal maturation and myelination of white matter in the developing brain. Am J Neuroradiol. 2009;30:290–296.
4. Song, S-K, Sun, S-W, Ramsbottom, MJ, et al. Dysmyelination revealed through MRI as increased radial (but unchanged axial) diffusion of water. Neuroimage. 2002;17(3):1429–1436.
5. Song, S-K, Sun, S-W, Ju, W-K, et al. Diffusion tensor imaging detects and differentiates axon and myelin degeneration in mouse optic nerve after retinal ischemia. Neuroimage. 2003;20:1714–1722.
6. Kim, JH, Loy, DN, Liang, H-F, et al. Noninvasive diffusion tensor imaging of evolving white matter pathology in a mouse model of acute spinal cord injury. Magn Reson Med. 2007;58:253–260.
7. Jeurissen, B, Leemans, A, Tournier, J-D, et al. Investigating the prevalence of complex fiber configurations in white matter tissue with diffusion magnetic resonance imaging. Hum Brain Mapp. 2012. [doi: 10.1002/hbm.22099 [E-pub ahead of print]].
8. Behrens, TEJ, Johansen-Berg, HJ, Jbabdi, S, et al. Probabilistic diffusion tractography with multiple fibre orientations: What can we gain? Neuroimage. 2007;34(1):144–155.
9. Mori, S, van Zijl, PCM. Fiber tracking: principles and strategies—a technical review. NMR Biomed. 2002;15:468–480.
10. Hirsch, JG, Schwenk, SM, Rossmanith, C, et al. Deviations from the diffusion tensor model as revealed by contour plot visualization using high angular resolution diffusion-weighted imaging (HARDI). MAGMA. 2003;16(2):93–102.
11. Tuch, DS. Q-ball imaging. Magn Reson Med. 2004;52(6):1358–1372.
12. Hess, CP, Mukherjee, P, Han, ET, et al. Q-ball reconstruction of multimodal fiber orientations using the spherical harmonic basis. Magn Reson Med. 2006;56(1):104–117.
13. Minati, L, Banasik, T, Brzezinski, J, et al. Elevating tensor rank increases anisotropy in brain areas associated with intra-voxel orientational heterogeneity (IVOH): a generalised DTI (GDTI) study. NMR Biomed. 2008;21(1):2–14.
14. Ozarslan, E, Shepherd, TM, Vemuri, BC, et al. Fast orientation mapping from HARDI. Med Image Comput Assist Interv. 2005;8(pt 1):156–163.
15. Ying, LL, Zou, YM, Klemer, DP, et al. Determination of fiber orientation in MRI diffusion tensor imaging based on higher-order tensor decomposition. Conf Proc IEEE Eng Med Biol Soc. 2007;2007:2065–2068.
16. Jensen, JH, Helpern, JA, Ramani, A, et al. Diffusional kurtosis imaging: the quantification of non-gaussian water diffusion by means of magnetic resonance imaging. Magn Reson Med. 2005;53(6):1432–1440.
17. Cheung, MM, Hui, ES, Chan, KC, et al. Does diffusion kurtosis imaging lead to better neural tissue characterization? A rodent brain maturation study. Neuroimage. 2009;45(2):386–392.
18. Raab, P, Hattingen, E, Franz, K, et al. Cerebral gliomas: diffusional kurtosis imaging analysis of microstructural differences. Radiology. 2010;254(3):876–881.
19. Basser, PJ. Relationships between diffusion tensor and q-space MRI. Magn Reson Med. 2002;47(2):392–397.

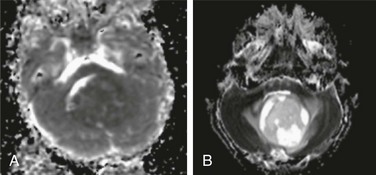
 , or axial diffusivity) being along the axis of preferred diffusion and the remaining two eigenvalues (λ2 and λ3) corresponding to vectors perpendicular to the principal axis. Eigenvalues λ2 and λ3 often are combined (averaged) to generate another metric referred to as radial diffusivity (λ
, or axial diffusivity) being along the axis of preferred diffusion and the remaining two eigenvalues (λ2 and λ3) corresponding to vectors perpendicular to the principal axis. Eigenvalues λ2 and λ3 often are combined (averaged) to generate another metric referred to as radial diffusivity (λ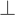 ). Importantly, although the 3 × 3 diffusion tensor may be ideal for estimating diffusion in a three-dimensional (3D) structure characterized by fiber bundles aligned in a single orientation, this model falls short if the underlying structure includes fiber bundles of different orientations (e.g., “crossing fibers”). Accordingly, potential pitfalls associated with DTI will be discussed in further detail later.
). Importantly, although the 3 × 3 diffusion tensor may be ideal for estimating diffusion in a three-dimensional (3D) structure characterized by fiber bundles aligned in a single orientation, this model falls short if the underlying structure includes fiber bundles of different orientations (e.g., “crossing fibers”). Accordingly, potential pitfalls associated with DTI will be discussed in further detail later.