Chryseobacterium, Sphingobacterium, and Similar Organisms
1. Describe the general characteristics of the organisms discussed in this chapter.
2. Identify the normal habitat and the routes of transmission for the organisms.
3. List the appropriate media for cultivation of the organisms listed, particularly E. meningoseptica.
4. Describe the colonial appearance of E. meningoseptica.
5. Outline the tests used to differentiate the major genera in this group, including Elizabethkingia sp., Myoides spp., Sphingobacterium spp., and Bergeyella zoohelicum.
General Characteristics
The organisms discussed in this chapter are environmental inhabitants that are occasionally encountered in human specimens. Most of the organisms originated in the heterogenous group Flavobacterium. However, when subjected to molecular analysis, they did not prove to be closely related and therefore have been reclassified. They are considered together here because they share similar physiologic and morphologic characteristics. Most are yellow-pigmented, oxidase-positive, glucose oxidizers that grow on MacConkey agar. Sphingobacterium spp. have an unusually large amount of sphingophospholipid compounds in their cell membranes. Sphingobacterium mizutaii, which does not grow on MacConkey agar, is discussed in Chapter 27.
Epidemiology
As environmental inhabitants, these organisms may be found in various niches (Table 24-1). Most notable in terms of clinical relevance is their ability to survive in hospital environments, especially in moist areas. Although they are not considered part of normal human flora, these species can colonize a patient’s respiratory tract during hospitalization. This results from exposure to contaminated water or medical devices. Transmission also may occur directly from contaminated pharmaceutical solutions and, in the case of E. meningoseptica, from person to person.
TABLE 24-1
| Species | Habitat (Reservoir) | Mode of Transmission |
| Elizabethkingia meningoseptica, Chryseobacterium spp., Empedobacter brevis, Sphingobacterium spp. | Soil, plants, water, food, and hospital water sources, including incubators, sinks, faucets, tap water, hemodialysis systems, saline solutions, and other pharmaceuticals Not part of human flora |
Exposure of patients to contaminated medical devices or solutions, but source is not always known. May colonize upper respiratory tract. E. meningoseptica occasionally may be transmitted from birth canal to neonate. |
| Chryseobacterium indologenes | Catheter-related infections | |
| Bergeyella zoohelicum | Normal oral flora of dogs and other animals | Dog and cat bites |
Pathogenesis and Spectrum of Disease
The development of infection basically requires exposure of debilitated patients to a contaminated source, resulting in respiratory colonization (Table 24-2). Depending on the patient’s health, subsequent infections, such as bacteremia and pneumonia, may develop. These infections are most frequently caused by Elizabethkingia meningoseptica or Myroides odoratus. Infections of several other body sites, which may or may not be preceded by respiratory colonization, have been associated with the other species.
TABLE 24-2
Pathogenesis and Spectrum of Diseases
| Species | Virulence Factors | Spectrum of Disease and Infections |
| Elizabethkingia meningoseptica, Chryseobacterium spp., Empedobacter brevis, Sphingobacterium spp. | Specific virulence factors are unknown. Able to survive chlorinated tap water. E. meningoseptica, the species most often associated with human infections, can be encapsulated or produce proteases and gelatinases that destroy host cells and tissues. |
Bacteremia (often associated with implanted devices, such as catheters, or contaminated medical solutions). E. meningoseptica is particularly associated with meningitis in neonates and less commonly in adults. Other organisms are associated with pneumonia, mixed infections of wounds, ocular and urinary tract infections, and occasionally sinusitis, endocarditis, peritonitis, and fasciitis. |
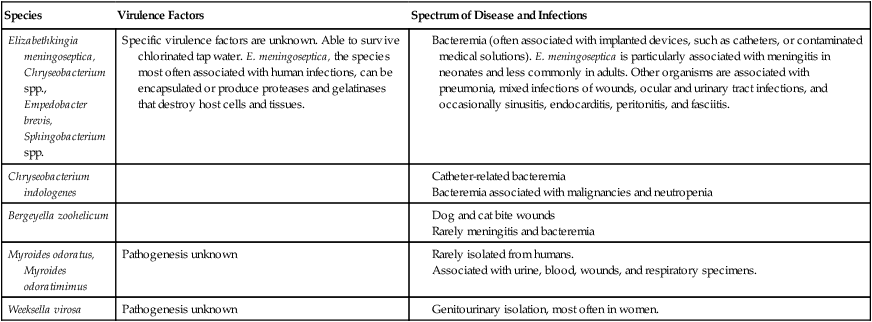
Meningitis caused by E. meningoseptica is the most notable infection associated with the organisms listed in Table 24-2. This life-threatening infection, which may be accompanied by bacteremia, originally gained attention because it occurred in neonates. However, E. meningoseptica meningitis can also occur in compromised adults. The organism has been implicated in hospital-based outbreaks of both meningitis and pneumonia.
Laboratory Diagnosis
Specimen Collection and Transport
No special considerations are required for specimen collection and transport of the organisms discussed in this chapter. Refer to Table 5-1 for general information on specimen collection and transport.
Cultivation
Media of Choice
Colonial Appearance
Table 24-3 presents descriptions of the colonial appearance and other distinguishing characteristics of each genus on 5% sheep blood and MacConkey agars.
TABLE 24-3
C. indologenes
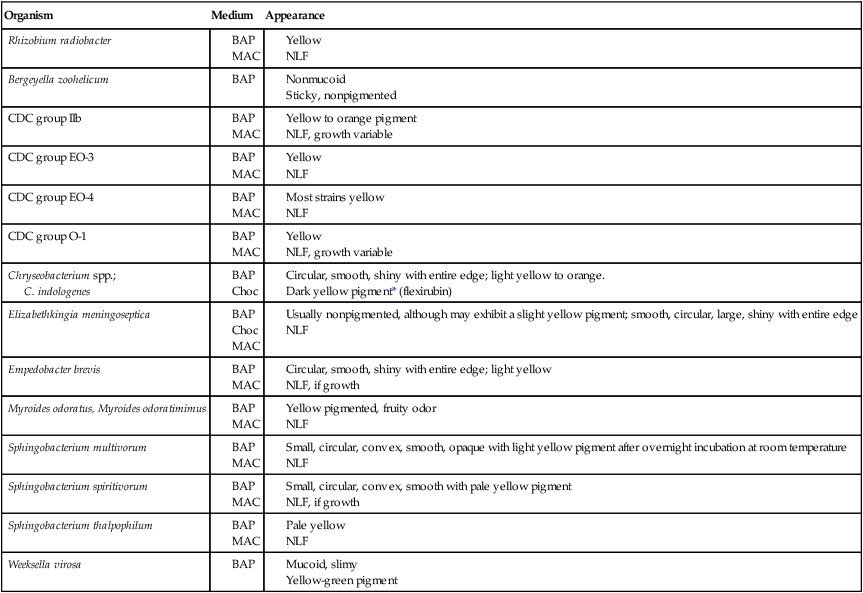
BA, 5% sheep blood agar; Mac, MacConkey agar; Choc, chocolate agar; NLF, non–lactose fermenter.
*Chryseobacterium spp. produce a yellow pigment that turns red upon the addition of 20% KOH.
Approach to Identification
The ability of most commercial identification systems to accurately identify the organisms discussed in this chapter is limited or uncertain. The key biochemical reactions used to presumptively differentiate among the genera discussed in this chapter are provided in Table 24-4. However, definitive identification of these organisms often requires a battery of biochemical tests not commonly available in many clinical microbiology laboratories. Therefore, full identification of clinically relevant isolates may require that they be sent to a reference laboratory.
TABLE 24-4
Key Biochemical and Physiologic Characteristics
| Organism | Oxidizes Mannitol | Indole | Gelatin | Urea | Nitrate Reduction | Esculin Hydrolysis | Motility |
| Agrobacterium yellow groupa | – | – | – | + | – | (+) | p,1-2 |
| CDC group EO–3 | (+) | – | – | (+) | – | – | nm |
| CDC group EO–4 | – | – | – | + | – | – | nm |
| CDC group O–1 | – | – | v | – | – | + | p, 1-2 |
| Chryseobacterium spp.b,d | – | + | v | v | v | v | nm |
| Elizabethkingia meningosepticab,c | + | + | + | – | – | + | nm |
| Empedobacter brevisb,c | – | + | + | – | – | – | nm |
| Myoides spp. | ND | _ | + | + | + | ND | nm |
| Sphingobacterium multivorum | – | – | – | + | – | + | nm |
| Sphingobacterium spiritivorum | + | – | v | + or (+) | – | + | nm |
| Sphingobacterium thalpophilum | – | – | v | + | + | + | nm |
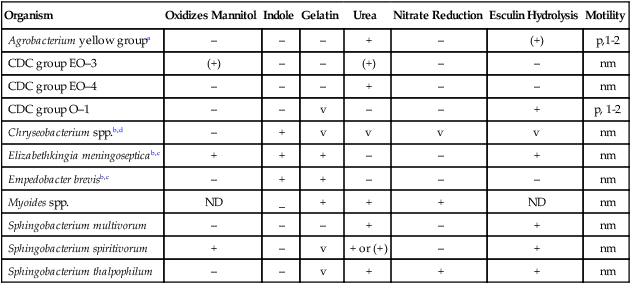
ND, No data; nm, nonmotile; p, polar flagella; v, variable; +, >90% of strains are positive; –, >90% of strains are negative; (+), reaction may be delayed.
aOnly a positive 3-ketolactonate test differentiates this group from Sphingomonas paucimobilis.
bColonial pigmentation is critical to separate Chryseobacterium spp. and Empedobacter brevis.
dIncludes Chryseobacterium gleum, C. indologenes, and CDC group IIb.
Comments Regarding Specific Organisms
The growth of Sphingobacterium spiritivorum and Chryseobacterium spp. is variable on MacConkey agar. Therefore, these organisms often need to be differentiated from the yellow-pigmented, MacConkey-negative, oxidase-positive genera considered in Chapters 27 and 31.
Antimicrobial Susceptibility Testing and Therapy
Validated susceptibility testing methods do not exist for these organisms. Although they grow on the media and under the conditions recommended for testing (see Chapter 12 for more information about validated testing methods), the ability to grow and the ability to detect important antimicrobial resistances are not the same. Therefore, the lack of validated in vitro susceptibility testing methods does not allow definitive treatment and testing guidelines to be given for any of the organisms listed in Table 24-5.
TABLE 24-5
Antimicrobial Therapy and Susceptibility Testing
| Species | Therapeutic Options | Potential Resistance to Therapeutic Options | Validated Testing Methods* | Comments |
| Bergeyella zoohelicum | Susceptible to penicillin | Not available | ||
| Chryseobacterium indologenes, Elizabethkingia meningoseptica, Empedobacter brevis, Sphingobacterium spp. | No definitive guidelines. Potentially active agents include ciprofloxacin rifampin, clindamycin, trimethoprim/ sulfamethoxazole, and vancomycin | Produce β-lactamases and are frequently resistant to aminoglycosides | Not available | In vitro susceptibility results with disk diffusion may be seriously misleading |
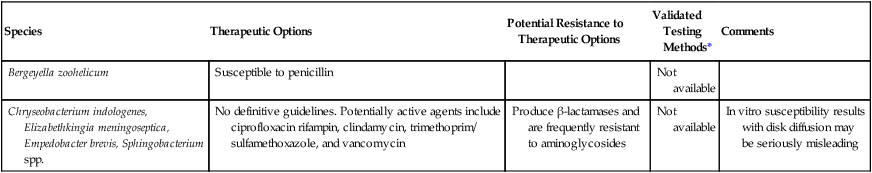
*Validated testing methods include standard methods recommended by the Clinical and Laboratory Standards Institute (CLSI) and commercial methods approved by the U.S. Food and Drug Administration (FDA).
Although susceptibility data for some of these bacteria can be found in the literature, the lack of understanding of potential underlying resistance mechanisms prohibits the validation of such data. Review Chapter 12 for preferable strategies that can be used to provide susceptibility information and data when validated testing methods do not exist for a clinically important bacterial isolate.







