CHAPTER 38 Cellular Motility
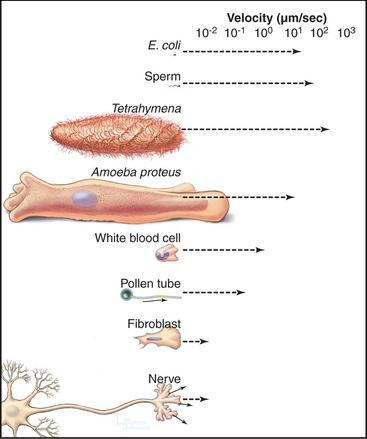
Cells move at rates that range over four orders of magnitude (Fig. 38-1 and Table 38-1). At one extreme, ciliates, bacteria, and sperm swim rapidly through water, and giant amoebas crawl rapidly over solid substrates. At the other extreme, fungal, algal, and plant cells with rigid cell walls are immobile. However, even some plant cells move, such as pollen, which extends tubular pseudopods. Most cells, including white blood cells, nerve growth cones, and fibroblasts move at intermediate rates.
Cells produce forces for motility in many different ways, most commonly using the same four mechanisms that produce intracellular movements (see Chapter 37): contraction of actin-myosin networks, movement of motors on microtubules, reversible assembly of actin filaments, or reversible assembly of microtubules. These mechanisms often complement each other, even where movement depends mainly on one system. For example, microtubules contribute to actin-based pseudopod extension by helping to specify the polarity of the cell. The chapter compares these standard mechanisms with a few novel mechanisms: contraction of calcium-sensitive fibers of ciliates, reversible assembly of novel cytoskeletal polymers of nematode sperm, and rotation of bacterial flagellar motors.
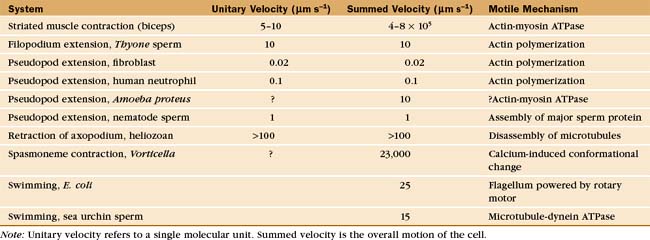
Most cells possess the proteins that are required for cellular motility, so the striking variation in their rates of movement arises from differences in the abundance and organization of this machinery. For example, both nonmotile yeasts and contractile muscle cells contain actin, myosin-II, heterodimeric capping protein, α-actinin, and tropomyosin. Yeasts use these proteins for cytokinesis (see Fig. 44-24), while muscle assembles high concentrations of similar proteins into sarcomeres (see Figs. 39-2 and 39-3) for powerful, fast contractions.
Cell Shape Changes Produced by Extension of Surface Processes
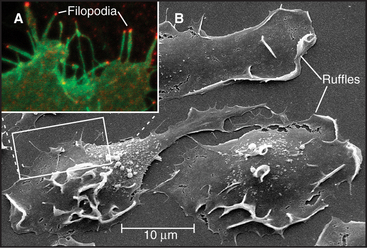
Simple alteration of cellular shape can be brought about by assembly of new cytoskeletal polymers or by rearrangement of preexisting assemblies of actin filaments or microtubules. One example that is dependent on assembly of actin filaments is the extension of cell surface projections called filopodia (Fig. 38-2).
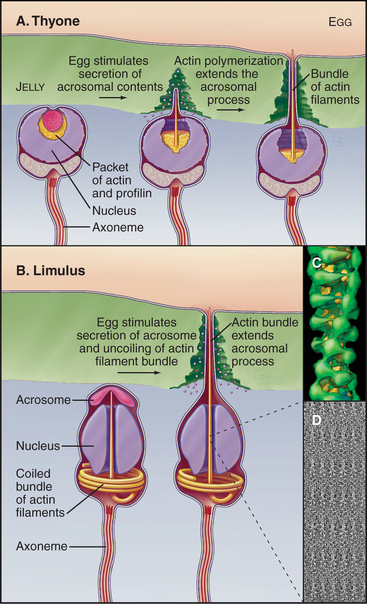
Studies of echinoderm sperm revealed that actin polymerization drives the formation of filopodia. Fertilization is accomplished when the sperm extend a long filopodium to penetrate the protective jelly surrounding the egg (Fig. 38-3A). Actin subunits for this acrosomal process are stored with profilin (see Figs. 1-4 and 33-19) in a concentrated packet near the nucleus. Contact with an egg stimulates actin filaments to polymerize, starting from a dense structure near the nucleus. Addition of subunits to the distal (barbed) end of growing filaments drives the elongation of the process and the surrounding membrane at a rate of 5 to 10 μm s−1 (an astounding maximum of 3700 subunits per second). Actin subunits diffuse rapidly enough from their storage site to drive this rapid elongation, which pushes the plasma membrane forward.
Filopodia on macrophages, nerve growth cones (Fig. 38-7), fibroblasts, and epithelial cells grow much more slowly and depend on formins at their tips (Fig. 38-2) to guide barbed-end assembly. Microvilli of the brush border of epithelial cells (see Fig. 33-2) are short, stable filopodia. The bundles of actin filaments supporting microvilli are cross-linked to each other (by fimbrin and villin) and to the plasma membrane by myosin-I. Synthesis of these accessory proteins during embryonic development triggers assembly of microvilli. Similarly, cells with few microvilli can be induced to make more simply by increasing the level of villin.
Sperm of the horseshoe crab, Limulus, use a novel acrosomal process to fertilize an egg (Fig. 38-3B). They preassemble a coiled bundle of actin filaments cross-linked by a protein called scruin. This bundle is a tightly coiled spring. An encounter with an egg stimulates rearrangement of the cross-links, causing the actin bundle to unwind. Uncoiling drives the bundle through a channel in the nucleus followed by extension of a process surrounded by plasma membrane that literally screws its way through the egg jelly to fuse with the egg plasma membrane.
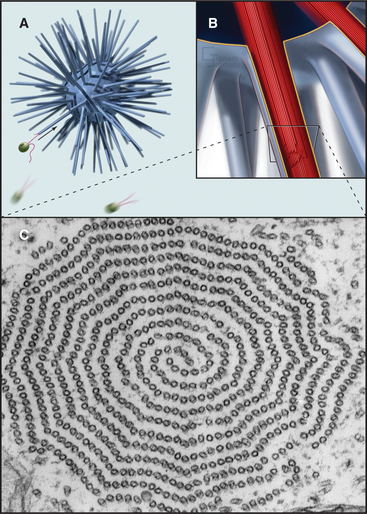
A group of ciliates called heliozoans, named for their similarity to a cartoon of the sun, are unique in using microtubules instead of actin filaments to extend, support, and retract long, thin processes bounded by the plasma membrane (Fig. 38-4). Microtubules in these axopodia are cross-linked into a precise geometrical array that accounts for the rigidity of these long processes. After mechanical stimulation by prey organisms, axopodia collapse in a few seconds, dragging the prey toward the cell body for phagocytosis. The collapse is caused by rapid depolymerization of the microtubules. Ca2+ influx appears to trigger depolymerization of the microtubules, but the details of the mechanism are not known.
Cell Shape Changes Produced by Contraction
Contraction by Actin and Myosin
Cells can change shape by localized or oriented cytoplasmic contractions. Muscle contraction (see Chapter 39) and cytokinesis (see Chapter 44) are the best examples, but contractions remodel many embryonic tissues. Localized contractions at the base or apex of cells in a planar epithelium cause evaginations or invaginations that form the neural tube and glands that bud from the gastrointestinal tract and respiratory tract (Fig. 38-5). Closure of the epidermis over a Drosophila embryo also requires contraction of a circumferential ring of cells. Tension generated by myosin-II and actin filaments deforms each cell and, collectively, the whole epithelium. Similarly, contraction of a ring of actin filaments associated with the zonula adherens of intestinal epithelial cells is one factor regulating the permeability of the tight junctions that seal sheets of epithelial cells (see Fig. 31-2).
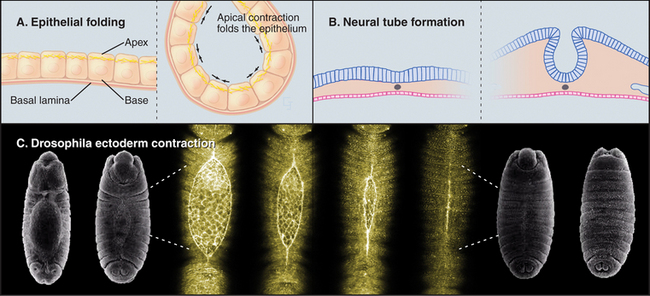
(C, SEMs courtesy of Thom Kaufman, Indiana University, Bloomington [see his movie “Fly Morph-o-genesis” at http://www.sdbonline.org/archive/dbcinema/kaufman/kaufman.html]; light micrographs courtesy of D. Kiehart, Duke University, Durham, North Carolina. Reference: Kiehart DP, Galbraith CG, Edwards KA, et al: Multiple forces contribute to cell sheet morphogenesis for dorsal closure in Drosophila. J Cell Biol 149:471–490, 2000.)
Calcium-Sensitive Contractile Fibers
The ciliate Vorticella avoids predators by contracting a stalk that anchors them to leaves or other supports (Fig. 38-6A). The contractile fibril, called a spasmoneme, contracts faster than any muscle. Ca2+ released from tubular membranes associated with the spasmoneme triggers contractions, when it binds to a calmodulin-like protein, spasmin, that forms 3-nm filaments in the spasmoneme. Ca2+ binding changes the conformation of spasmin and results in rapid shortening, be-cause many spasmin subunits are assembled in series. The spasmoneme relaxes when Ca2+ dissociates. Energy for contraction is supplied indirectly by ATP hydrolysis. ATP-driven pumps create a Ca2+ gradient between the lumen of the membrane system and cytoplasm. Movement of Ca2+ down this gradient drives contraction.
Proteins similar to spasmin are found not only in other ciliates but also in algae, fungi, and animals, where they are called centrin or caltractin. These calmodulin-like proteins form fibrils that anchor centrosomes and the basal bodies of cilia and flagella. Mutations that inactivate caltractin in algae or yeast compromise the duplication and separation of the microtubule organizers (centrosomes or spindle pole bodies; see Figs. 34-16 and 34-19) used for mitosis.
Locomotion by Pseudopod Extension
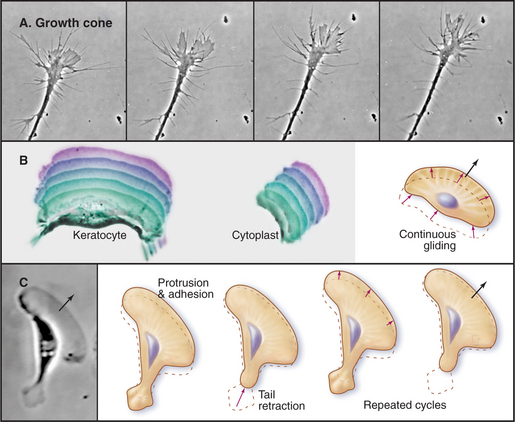
The ability to crawl over solid substrates or through extracellular matrix is essential for many cells. Perhaps the most spectacular example is the slowly moving growth cone of a nerve axon (Fig. 38-7A). Although moving less than 50 μm s−1, growth cones navigate precisely over distances ranging from micrometers to meters to establish all of the connections in the human nervous system, which consists of billions of neurons and about 1 million miles of cellular processes. Some epithelial cells (Fig. 38-7B) and white blood cells move much faster, about 0.5 μm s−1. These movements enable epithelial cells to cover wounds and allow leukocytes to move from the blood circulation to sites of inflamma-tion (see Fig. 30-13) and to engulf microorganisms by phagocytosis (see Fig. 22-3). During vertebrate embryogenesis, neural crest cells also migrate long distances before differentiating into pigment cells and sympathetic neurons. Fibroblasts lay down collagen fibrils as they move through the extracellular matrix (see Fig. 29-4).
This type of locomotion requires coordination of three different events. The cell must extend its leading edge, adhere to the underlying substrate, and (if the whole cell is to move) retract any attachments of its tail to the substrate. Most cells use assembly of actin filaments to extend pseudopods, but nematode sperm accomplish the same thing with a completely different protein (Fig. 38-11).
Pseudopod Extension
Pseudopods that lead the way in cell migration are filled with a dense, branched network of actin filaments with their fast-growing barbed ends generally facing the plasma membrane (Fig. 38-8). Only the leading lamellum is required for locomotion, since it moves normally after amputation from the rest of the cell (Fig. 38-7B). Generally, the leading lamellum is very flat, on the order of 0.25 μm thick, but some cells extend the lamellum up from the substrate into a wave-like fold of membrane called a ruffle (Fig. 38-2). Microtubules help cells to maintain the polarized shape that is required for persistent directional locomotion, but they are not required for pseudopod extension. A role for actin polymerization in pseudopod extension was originally indicated by the ability of the drug cytochalasin to inhibit the process (see Fig. 33-18).
(Redrawn from Pollard TD, Blanchoin L, Mullins RD: Biophysics of actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct 29:545–576, 2000, with permission from the Annual Review of Biophysics and Biomolecular Structure, Volume 29, © 2000 by Annual Reviews, www.annualreviews.org. Inset, Courtesy of T. Svitkina and G. Borisy, Northwestern University, Evanston, Illinois.)
Microscopic observations of cells injected with fluorescent actin molecules show that filaments assemble continuously near the leading edge of pseudopods (Fig. 38-9). Purified actin can be labeled with a fluorescent dye and microinjected into live cells, where it incorporates into actin-containing structures, including the cortical network, pseudopods, stress fibers, and surface microspikes. If the dye bound to actin is bleached locally with a strong pulse of light inside a stationary cell (Fig. 38-9A), the bleached spot moves away from the edge of the cell. The spot recovers as bleached actin is replaced with fluorescent actin through a combination of diffusion and active movement of filaments, assembly of new filaments, or subunit flux through filaments. To assess the contribution of each process, a cell can be injected with actin carrying a “caged” dye. The caged dye is not fluorescent until a blocking group is removed locally by photolysis with a pulse of light (Fig. 38-9B). Fluorescent actin monomers diffuse away quickly, so any fluorescent filaments can be observed. In rapidly moving cells, marked filaments remain relatively stationary with respect to the substrate as the front of the cell advances, confirming that new filaments are assembled at the leading edge. The fluorescence of the marked filaments declines over a period of minutes, as fluorescent subunits are released from filaments and diffuse away. In a third approach, a low concentration of fluorescent actin is injected such that it incorporates irregularly into filaments, producing spots of fluorescence that can be tracked over time (see Fig. 33-18). Analysis of these fluorescent speckles confirms that filaments assemble at the leading edge and turn over rapidly deeper in the cytoplasm.
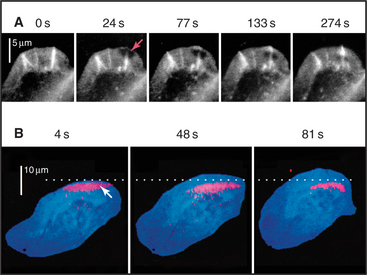
Figure 38-9 documentation of actin filament dynamics at the leading edge with fluorescent actins. A, Fluorescence photobleaching experiment with a stationary cell. Fluorescent actin is injected into a cultured epithelial cell and allowed to incorporate into filaments. A laser pulse bleaches some of the fluorescent actin, leaving a dark spot (arrow) that reveals movement of the filaments toward the cell center. B, Caged fluorescent actin experiment with a motile cell. Fluorescent dye bound to actin is masked with a chemical group preventing fluorescence. After incorporation into actin filaments of a fish keratocyte (see Fig. 33-2E), dyes in one area of the cell are uncaged with a light pulse (arrow), and red fluorescence is followed with time. Fluorescent actin filaments are stationary with respect to the substrate as the cell moves forward (upward). The fluorescent spot of marked filaments fades with time, owing to depolymerization and dispersal of the fluorescent subunits.
(A, Reproduced from Wang Y-L: Exchange of actin subunits at the leading edge of living fibroblasts: Possible role of treadmilling. J Cell Biol 101:597–602, 1985, by copyright permission of The Rockefeller University Press. B, From Theriot JA, Mitchison TJ: Actin microfilament dynamics in locomoting cells. Nature 352:126–131, 1991.)
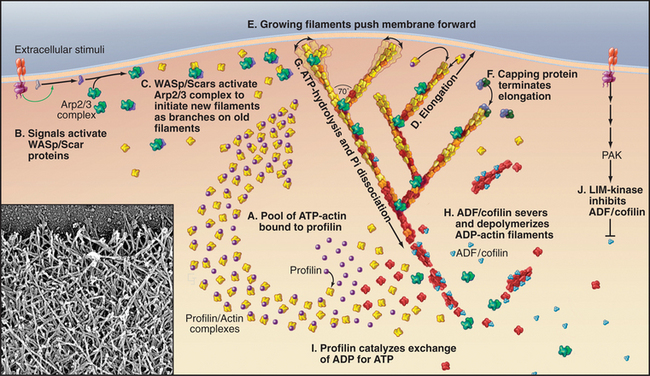
The molecular mechanism that assembles actin filaments at the leading edge (Fig. 38-8) shares many features with the formation of actin filament comet tails by intracellular bacteria (see Fig. 37-12) and by actin patches in yeast (Figs. 6-3 and 37-11). Chemotactic stimuli (Fig. 38-12) or intrinsic signals transduced by Rho-family GTPases, membrane polyphosphoinositides, and proteins with SH3 domains activate WASp/Scar proteins, which promote the formation of actin filament branches by Arp2/3 complex (see Fig. 33-13). The pool of unpolymerized actin maintained by profilin (and thymosin-b4, where it is present) drives the elongation of actin filament branches at 50 to 500 subunits per second. The growing filaments are generally oriented toward the leading edge and push against the inside of the plasma membrane with forces in the piconewton range. Heterodimeric capping protein terminates elongation of the branches before they grow longer than 1μm. Longer filaments are less effective at pushing, since they buckle under piconewton forces.
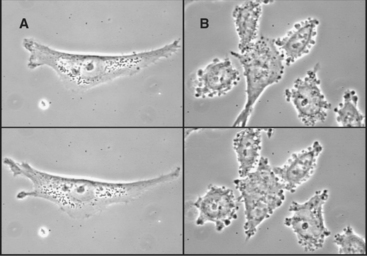
Actin filament cross-linking proteins stabilize pseudopods. Human melanoma cells that lack filamin form unstable pseudopods all around their peripheries and locomote abnormally (Fig. 38-10). These tumor cells recover their normal behavior when provided with filamin. Similarly, Dictyostelium cells that lack a homolog of filamin form fewer pseudopods.
The recycling of actin and accessory proteins is essential for multiple rounds of assembly as the cell moves forward. Severing proteins such as ADF/cofilins are thought to promote the disassembly of aged ADP-actin filaments located away from the leading edge, although the details are not established. One mystery is how the branched network is rapidly converted into long, unbranched filaments a short distance behind the leading edge (see Fig. 33-2D-E). The side-binding protein tropomyosin protects these longer filaments from ADF/cofilins.
Adhesion: Influence of the Substrate
Pseudopods must establish contacts with the substrate for a cell to move forward. Cells tend to move up gradients of adhesiveness but stop if adhesion is too strong, so adhesion with movement requires a compromise. Adhesion must be strong enough for the internal forces to propel the cell forward but not so strong that it prevents movement. Rapidly reversible binding of integrins and other adhesion proteins to extracellular matrix molecules, such as fibronectin, allows adhesion without immobilization. Rapidly moving white blood cells attach weakly and transiently, whereas slowly moving fibroblasts form longer-lasting focal contacts (see Fig. 30-11). Both the chemical nature and the physical nature of the substrate influence adhesion and movement. Cultured cells move up gradients of fibronectin coated on glass. Similarly, neural crest cells migrate preferentially through regions of embryonic connective tissue marked by adhesive proteins.
The growing actin network at the leading edge will either push the membrane forward or slip backward depending on how well it is connected to the substrate across the plasma membrane. In highly motile cells such as epithelial cells from fish scales (Fig. 38-7B) transmembrane adhesion proteins anchor the actin filament network to the substrate, so the polymerization results in forward motion. In stationary cells (Fig. 38-9A), actin polymerizes at the edge of the cell but the entire network moves en masse away from the membrane, a phenomenon called retrograde flow. Fibroblasts are an intermediate state, in which actin polymerization produces some forward movement in addition to considerable retrograde flow. Another variation of this theme is seen in nerve growth cones (Fig. 38-7A), which extend short filopodia and then fill the spaces in between with a lamellum filled with new actin filaments.
Tail Retraction and Other Roles for Myosin in Motility
Growth cones draw out a long process from a stationary cell body, but most cells must break adhesions at their trailing edge to advance. Adherent, slowly moving cells such as fibroblasts exert significant tension on the underlying substrate, when myosin pulls on the actin filaments associated with focal contacts. When tension overcomes the attachments, the rear of the cell shortens elastically and then contracts further (Fig. 38-7C). Myosin also contributes to the retrograde flow of actin filaments in the zone between the leading edge and the cell body. Super-fast giant amoebas (Fig. 38-1) appear to use myosin to generate contractions in the cortex or the front of the pseudopod to drive the bulk streaming of cytoplasm into advancing pseudopods.
An Actin Substitute in Nematode Sperm
Nematode sperm use amoeboid movements to find an egg rather than swimming with flagella like other sperm (Fig. 38-11). The behavior of these sperm is so similar to a small amoeba cell that anyone would have guessed that it is based on the assembly of actin filaments. However, actin is a minor protein in nematode sperm. Instead, sperm pseudopods are filled with 10-nm filaments assembled from a 14-kD protein called major sperm protein. Proteins in the cytoplasm and associated with the plasma membrane guide the assembly of the filaments, which function remarkably like actin, despite the fact that they have no bound nucleotide and no known associated motor protein. Light microscopy of migrating cells shows that 10-nm filaments assemble at the leading edge of the pseudopod and remain stationary with respect to the substrate as the expanding pseudopod advances. Filament bundles depolymerize at the interface between the pseudopod and the spherical cell body. A pH gradient is thought to influence assembly at the front and disassembly at the rear of the pseudopod. This highly efficient motility system is still unknown in other parts of the phylogenetic tree.
Chemotaxis of Motile Cells
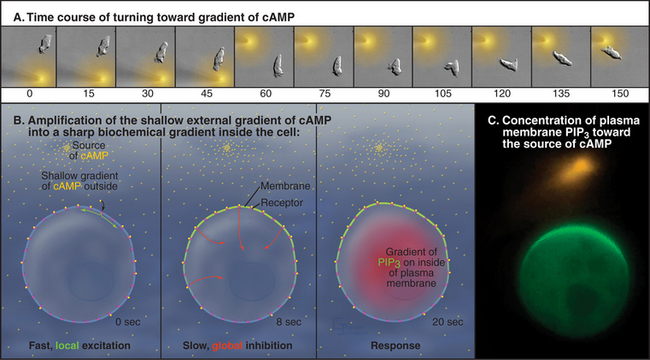
Extracellular chemical clues direct locomotion by influencing the formation and persistence of pseudopods. Movement toward a positive signal is called chemotaxis. The best-characterized example is the attraction of Dictyostelium to cAMP (Fig. 38-12), the extracellular chemical that these amoebas use to communicate as they form colonies before making spores. Remarkably, these cells can sense a gradient of cAMP corresponding to a concentration difference of less than 2% along their length. This small difference is amplified into strong internal signals that control motility. Binding of cAMP to seven-helix receptors in the plasma membrane activates trimeric G-proteins inside the cell (see Fig. 25-9). The G-proteins activate pathways that regulate the activity of enzymes that control the concentration of the lipid second messenger phosphatidylinositol 3,4,5 trisphosphate, PIP3, in the plasma membrane: phosphatidylinositol 3-kinase (PI3K) synthesizes and PTEN phosphatase degrades PIP3. (See Fig. 26-7 for details on polyphosphoinositides.) A fast positive pathway that is sensitive to local receptor occupancy and a slower global negative signal that is proportional to total receptor occupancy produce a gradient of PI3K activity inside the cell that is steeper than the external gradient of cAMP. These pathways have the opposite effect on PTEN, concentrating it on the plasma membrane away from the source of cAMP. This complementary regulation of the kinase and phosphatase creates an internal gradient of PIP3 three to seven times steeper than the external gradient of cAMP. Transduction of this internal gradient of PIP3 into motility requires Rho-family GTPases and formation of new actin filaments. Local polymerization and cross-linking of these actin filaments expand the cortex facing the source of cAMP into a new pseudopod and move the cell toward the cAMP.
Leukocytes are attracted to chemokines and bacteri-al metabolites at sites of infection (see Fig. 30-13), es-pecially small peptides from the N-termini of bacter-ial proteins, such as N -formyl-methionine-leucine-phenylalanine (referred to as FMLP in the scientific literature). Activation of seven-helix receptors and trimeric G-proteins amplifies shallow external gradients of FMLP into steeper internal gradients of PIP3 and other signals that control pseudopod formation.
Growth Cone Guidance: A Model for Regulation of Motility
Growth cones of embryonic nerve cells use a combination of positive and negative cues to navigate with high reliability to precisely the right location to create a synapse (Fig. 38-13). This combinatorial strategy is much more complex than the simple chemoattraction of Dictyostelium to cAMP, as expected for the more complicated task of connecting billions of neurons to each other and to targets, such as specific muscle cells. Cues for growth cone guidance come from soluble factors and cell surface molecules, each requiring a specific receptor on the growth cone. As in other systems, extracellular matrix molecules provide a substrate for growth cone movements. Precisely positioned expression of cue molecules and their receptors guides growth cones along a staggering number of different pathways. The following are some well-characterized examples.
Cell Adhesion Proteins
Ig-CAM cell surface adhesion proteins (see Fig. 30-3), such as fasciculin II, prompt growing axons to bundle together in bundles called fascicles by homophilic interactions. Growth cones can be attracted out of these bundles to particular targets, such as muscle cells, that secrete chemoattractants or proteins that antagonize fasciculin II adhesion.
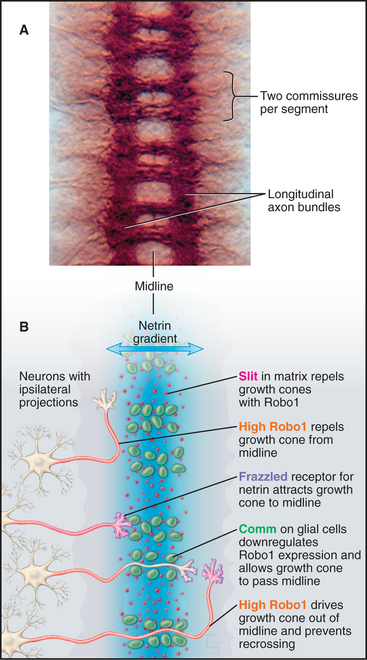
Navigation of growth cones in Drosophila embryos illustrates these multiple guidance cues (Fig. 38-13). The original insights came largely from studying the effects of mutations in genes for the various receptors and their ligands. Growth cones of neurons on one side of the nerve cord migrate across the midline to the opposite side and then navigate faithfully to their targets. Netrins secreted by cells at the midline attract growth cones expressing the netrin receptor. However, midline cells also secrete high levels of the matrix protein Slit, which repels growth cones. Growth cones cross the midline by downregulating the slit receptor. Once growth cones complete their journey across the midline, they upregulate the slit receptor, so they never cross back to the side of origin. Local cues alert particular growth cones of motor neurons to branch off of fascicles to innervate individual muscle cells. Path finding by capillaries uses some of the same guidance mechanisms to grow blood vessels.
Locomotion by Cilia and Flagella
Microtubule-containing axonemes that produce the beating of cilia and flagella are not only exceedingly complex but also remarkably ancient. Diplomonads that branched early in the eukaryotic radiation (see Fig. 2-4) have flagella that share the essential features of human cilia and flagella. This highly efficient organelle for rapid swimming developed well over a billion years ago and is retained essentially unchanged in many parts of the eukaryotic phylogenetic tree. Most protists, algae, and animals have axonemes, but most fungi, ferns, and plants have lost the genes for axonemes.
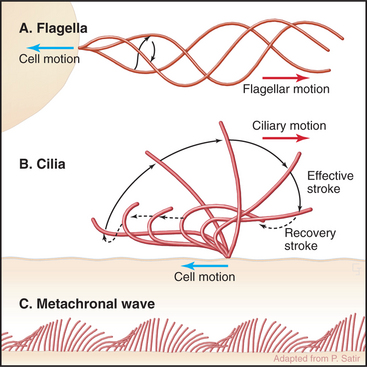
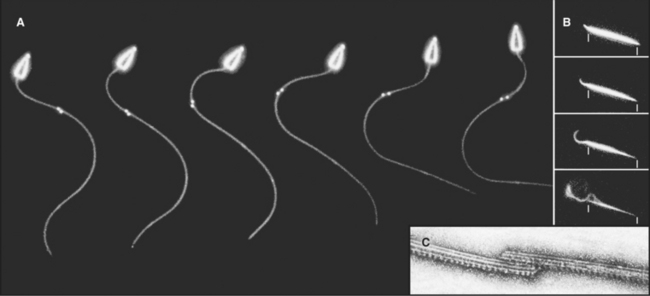
Cilia and flagella are distinguished from each other by their beating patterns (Fig. 38-14), but are nearly identical in structure. In fact, the flagella of the green alga Chlamydomonas can alternate between propagating waves typical of flagella and the oar-like rowing motion of cilia. Subtle differences in the mechanism that converts the dynein-powered sliding of the axonemal microtubules into movements determine which beating pattern is produced.
Both cilia and flagella can propel cells as they cycle rapidly, beating up to 100 times per second. Propagation of bends along the length of individual flagella pushes the cell forward. Coordinated beating of many cilia can move large cells (Fig. 38-15). Reversal of the direction of the power stroke allows a cell to swim forward or backward. Alternatively, if the cell is immobilized, like epithelial cells lining an animal respiratory tract, coordinated beating of cilia propels fluid and particles over their apical surface. Ctenophores fuse the membranes of many cilia together to make macrocilia that propel the organism in a manner similar to that of fins.
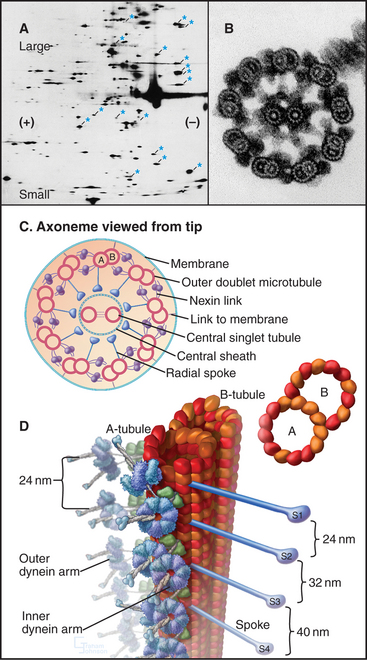
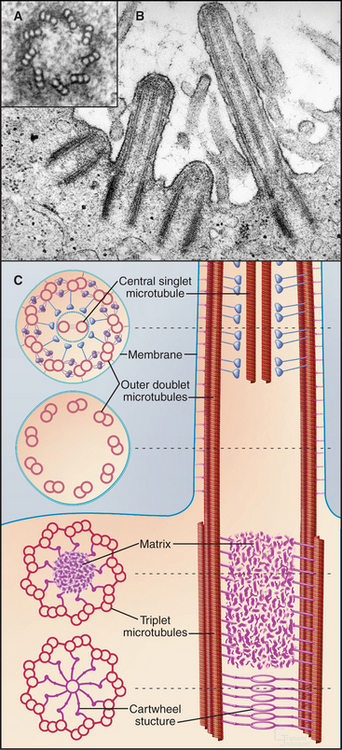
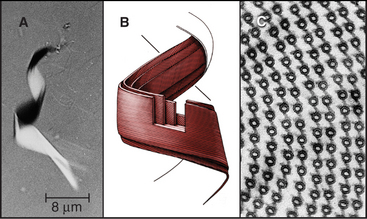
Although nature has produced some fascinating variations, most cilia and flagella consist of axonemes composed of a 9 + 2 arrangement of microtubules surrounded by the plasma membrane (Figs. 38-16 and 38-17). The 9 outer doublets consist of one complete α-microtubule of the usual 13 protofilaments, with an incomplete β-microtubule composed of 10 protofilaments attached to its side. Tektin, a filamentous protein in the wall of the α-microtubule, might help to attach the β-microtubule. Like most microtubular structures, the distal end of axonemal microtubules is the plus end. The central pair are typical 13-protofilament microtubules.
More than 200 accessory proteins reinforce the 9 + 2 microtubules (Fig. 38-16A), making axonemes stiff but elastic. Genetic analysis established the locations of many of these polypeptides, such as the 17 proteins that make up the radial spokes between the central sheath and the outer doublets. Circumferential links join outer doublets to each other. Central pair microtubules are connected by a bridge and decorated by elaborate projections.
A family of axonemal dyneins bound to outer doublets generates force for movement. Each dynein consists of a large heavy chain with globular AAA ATPase domain and a flexible stem anchored to an α-tubule by light and intermediate chains. A thin stalk projecting from the catalytic domain exerts force on the adjacent β-tubule during part of the ATPase cycle (see Figs. 36-14 and 36-15). The outer row of dynein arms in Chlamydomonas axonemes are all the same three-headed molecules (Fig. 38-16D). Seven different inner-arm dyneins are arranged in an orderly pattern that repeats every 96 nm along the α-tubule of each outer doublet.
Dynein-powered sliding of outer doublets relative to each other bends axonemes. Sliding was first inferred from electron micrographs of the distal tips of microtubules in bent cilia. Later, sliding was observed directly by loosening connections between outer doublets with proteolytic enzymes and then adding ATP to allow dynein to push the microtubules past each other (Fig. 38-18B). Sliding can be followed precisely in axonemes stripped of their membrane by marking outer doublets with small gold beads (Fig. 38-18A). As outer doublets slide past each other, the relative positions of the beads change. Dynein attached to one doublet “walks” toward the base of the adjacent microtubule, pushing its neighbor toward the tip of the axoneme.
The mechanism of beating is intrinsic to the axoneme, as sperm tail axonemes swim normally when provided with ATP, even without the plasma membrane or soluble cytoplasmic components (Fig. 38-18A). Experiments with these demembranated sperm models revealed that the dynein adenosine triphosphatase (ATPase) activity is tightly coupled to movement. The beat frequency is proportional to ATPase activity, regardless of whether the frequency is limited by increasing the viscosity of the medium or the enzyme activity is limited by decreasing the ATP concentration.
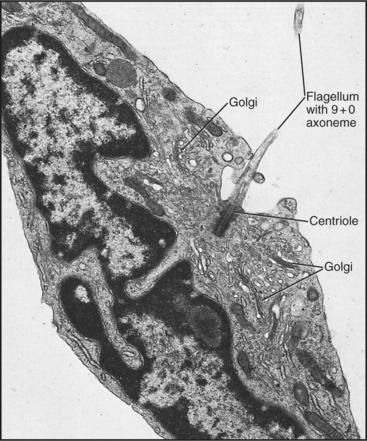
A basal body, a modified centriole similar to those in the centrosome of animal cells, anchors each axoneme in the cortex of the cell (Fig. 38-17; see also 34-3B). Note that the nine outer doublets of the axoneme grow directly from an extension of the nine outer triplet microtubules of the basal body rather than from amorphous pericentriolar material that initiates interphase microtubules (see Fig. 34-16). In sperm, one centriole serves as the basal body. In some protozoa, basal bodies are used as centrioles during mitosis. In ciliated cells, basal bodies replicate simultaneously from amorphous filamentous material to provide a basal body for each of the numerous axonemes.
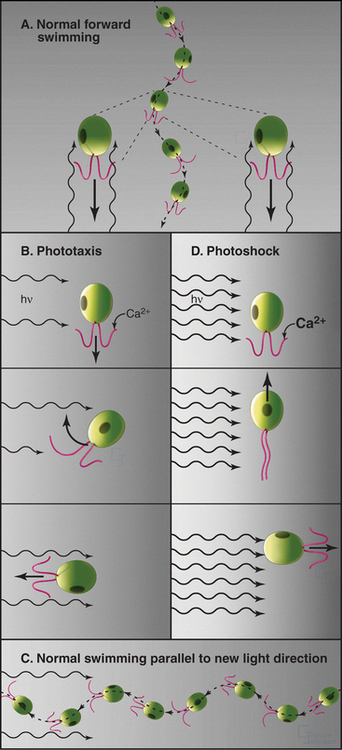
Although axonemes function autonomously, they are regulated by signal transduction pathways. Phototaxis of Chlamydomonas is a particularly clear example of how fluctuations in intracellular Ca2+ can modify flagellar activity. The release of Ca2+ affects the two flagella of the organism differentially and allows a cell to steer toward or away from light (Fig. 38-19). Ciliates also have mechanosensitive channels that depolarize the plasma membrane when the organism collides with something. Depolarization opens voltage-sensitive plasma membrane Ca2+ channels, admitting Ca2+ into the cell. This reverses the direction of ciliary beat. Both calcium and cAMP-dependent phosphorylation of outer-arm dynein can change the beat frequency (all the way to zero) or alter the wave form.
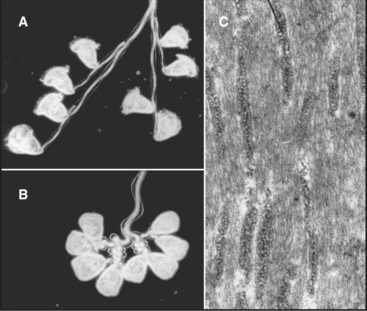
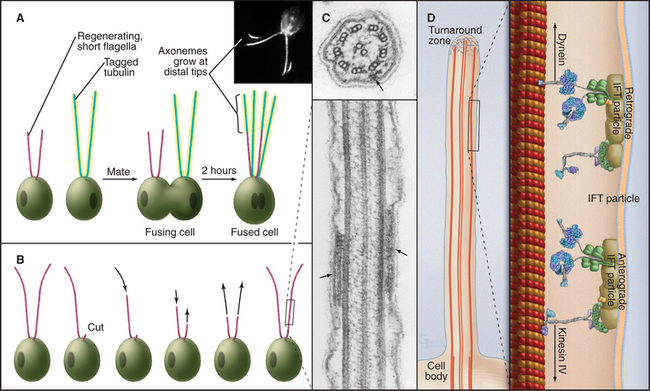
Some species regenerate flagella if they are severed from the cell (Fig. 38-20A-B). Absence of the flagellum activates expression of genes required to supply subunits for regrowth of the axoneme. In about 1 hour, the cell regrows a replacement flagella, and the genes are turned off. Even more remarkably, if only one of the two flagella is lost, the remaining flagellum shortens rapidly to provide components required to make two half-length flagella. Then protein synthesis slowly provides additional subunits to restore both flagella to full length.
Axonemes grow at their tips by incorporation of subunits synthesized in the cytoplasm. A process called intraflagellar transport (Fig. 38-20C-D) carries individual proteins and subassemblies such as radial spokes to the growing tip. Kinesin-2 motors move the packets of proteins toward the tip of the axoneme along the outer doublets just beneath the plasma membrane. Cytoplasmic dynein 1b transports particles back toward the cell body. Transmembrane proteins of the plasma membrane also move bidirectionally along microtubules of the underlying axoneme, presumably powered by motor proteins. Intraflagellar transport is remarkably similar to fast axonal transport (see Fig. 37-1) but on a smaller scale. In neither case is it known how transport is reversed for the return trip from the tips of the microtubules.
Primary Cilia
With the exception of blood cells, most differentiated cells in metazoan tissues produce a single primary cilium by growth of an axoneme from their older centriole (Fig. 38-21). The axonemes lack the central pair, and most lack dynein, so they are immotile. Many primary cilia are sensory organelles with special receptors in the plasma membrane. Nematode olfactory neurons have their odorant receptors concentrated in the membranes of primary cilia. Rod and cone photoreceptors in the eye are modified cilia with a basal body and a vestigial axoneme (see Fig. 27-2). Primary cilia on the epithelial cells of kidney tubules act as flow sensors, admitting Ca2+ into the cell through mechanosensitive channels in the plasma membrane when bent. Deficiencies in either intraflagellar transport or these ion channels result in polycystic kidney disease, a relatively common cause of kidney failure.
Specialized Microtubular Organelles
Some protozoa use dynein to generate beating movements of large arrays of cytoplasmic microtubules called axostyles (Fig. 38-22). The mechanism seems to be similar to an axoneme, although the organization clearly differs. Cross-linking structures hold together sheets of singlet microtubules, which slide past each other as a result of the action of dynein motors on adjacent sheets. Coordinated beats of the axostyle distort the whole organism, allowing it to wiggle about.
Bacterial Flagella
Bacteria use a reversible, high-speed, rotary motor driven by H+ or Na+ gradients to power their flagella (Figs. 38-23 and 38-24). Bacterial flagella differ in every respect from eukaryotic cilia and flagella. The bacterial flagellum is an extracellular protein wire (see Fig. 5-9), not a cytoskeletal structure like an axoneme inside the plasma membrane. Bacteria with multiple flagella are more common than those with single flagella.
(Redrawn from Schuster SD, Khan S: The bacterial flagellar motor. Annu Rev Biophys Biomol Struct 23:509–539, 1994, with permission from the Annual Review of Biophysics and Biomolecular Structure, Volume 23, © 1994 by Annual Reviews, www.annualreviews.org.)
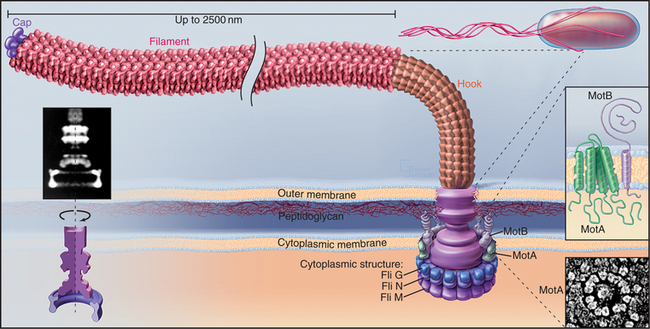
(Left, Courtesy of D. DeRosier, Brandeis University, Waltham, Massachusetts. Reference: Thomas DR, Morgan DG, DeRosier DJ: Rotational symmetry of the C ring. Proc Natl Acad Sci U S A 96:10134–10139, 1999. Middle, Redrawn from Schuster SD, Khan S: The bacterial flagellar motor. Annu Rev Biophys Biomol Struct 23:509–539, 1994, with permission from the Annual Review of Biophysics and Biomolecular Structure, Volume 23, © 1994 by Annual Reviews, www.annualreviews.org. Lower right, Courtesy of S. Khan, Albert Einstein College of Medicine, Bronx, New York.)
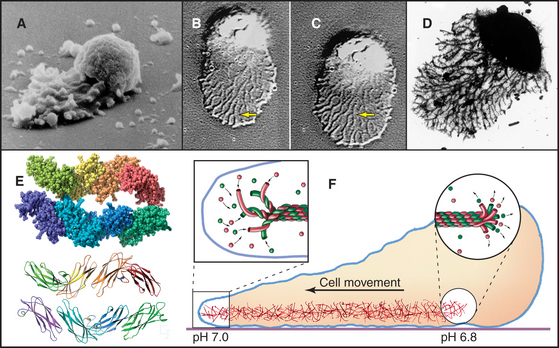
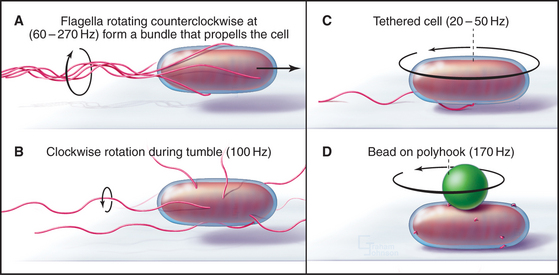
A motor, embedded in the plasma membrane, turns the bacterial flagellum either clockwise or counterclockwise (viewed from the tip of the flagellum) like the propeller of a motor boat. When multiple flagella are present, counterclockwise rotation forms a bundle. Four flagella propel Escherichia coli 30 μm s−1, a velocity of 15 cell lengths per second, equivalent to 400 miles per hour if the bacterium were the size of an automobile. When one or more flagella reverse their direction and rotate clockwise, the bundle flies apart, and the cell tumbles in one place. Figures 27-12 and 27-13 explain how chemotactic stimuli control the probability of clockwise rotation, favoring steady runs toward nutrients and allowing for more frequent tumbles to change direction to avoid harm.
Assays for rotation of single flagella provide insights about the mechanism of flagellar motion (Fig. 38-23). When a flagellum is attached to a glass slide by means of antibodies to the flagellar filament, the bacterium rotates, providing decisive evidence for rotation of flagella. Similarly, beads attached to short flagella are observed to rotate. The rotational speed depends on the resistance. The motor of a single immobilized flagellum can rotate a whole E. coli 10 to 50 times per second, whereas in some species, unloaded motors rotate up to 1600 times per second (100,000 rpm)!
The rotary engine driving the flagellar filament is constructed from two parts: a rotating, cylindrical basal body on the end of the filament and a surrounding ring of stationary proteins embedded in the plasma membrane and anchored to the peptidoglycan layer (Fig. 38-24). Genetic screens for motility mutants identified all of the protein components of the motor, and their functions were defined by analysis of the behavior of these mutants. Most of these proteins are present in isolated basal bodies. Two proteins essential for rotation—MotA and MotB—are found in the cell membrane surrounding the basal body. MotA has four hydrophobic segments that are believed to be transmembrane helices. MotB has transmembrane segment in addition to a periplasmic domain anchored to the peptidoglycan layer (Fig. 38-24). Flagella are immotile in cells that lack either one of these proteins. If the missing protein is replaced by initiating its biosynthesis, the paralyzed flagellum begins to turn, increasing its speed of rotation in a stepwise fashion, as independent, torque-producing units are added one after another. At the same time, the ring of 10 to 12 transmembrane proteins reappears surrounding the basal body, so these particles are believed to be the motors.
The energy to turn the motor comes from protons (or, in some bacteria, Na+) that move down an electrochemical gradient from outside the bacterium through the motor to the cytoplasm. Transfer of one proton across the membrane provides approximately the same energy as the hydrolysis of an ATP. Pumps driven by light, oxidation, or ATP hydrolysis (see Table 8-1) generate the proton gradient. Flagellar rotation stops when bacteria are starved, and it resumes when nutrients are supplied to allow reestablishment of the membrane proton gradient. MotA is the prime candidate for the proton channel, because mutations in its gene inhibit both flagellar rotation and proton permeability. Roughly 1000 protons cross the membrane for each rotation, corresponding to two protons for each tiny rotational step. Proton transfer is tightly coupled to rotation of the basal body, and the efficiency is near 100%. Detailed understanding of the mechanism awaits determination of atomic structures of the proteins.
Beisson J, Wright M. Basal body/centriole assembly and continuity. Curr Opin Cell Biol. 2003;15:96-104.
Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19-54.
Bray D. Cell Movements, 2nd ed. New York: Garland Publishing, 2000.
Carmeliet P, Tessier-Lavigne M. Common mechanisms of nerve and blood vessel wiring. Nature. 2005;436:193-2000.
Condeelis J, Singer RH, Segall JE. The great escape: When cancer cells hijack the genes for chemotaxis and motility. Annu Rev Cell Dev Biol. 2005;21:695-718.
Huber AB, Kolodkin AL, Ginty DD, Cloutier JF. Signaling at the growth cone, ligand-receptor complexes and the control of axon growth and guidance. Annu Rev Neurosci. 2003;26:509-563.
Jaffe AB, Hall A. Rho GTPases: Biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247-269.
Kamiya R. Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int Rev Cytol. 2002;219:115-155.
Levin M. Left-right asymmetry in embryonic development: A comprehensive review. Mech Dev. 2005;122:3-25.
Manahan CL, Iglesias PA, Long Y, Devreotes PN. Chemoattractant signaling in Dictyostelium discoideum. Annu Rev Cell Dev Biol. 2004;20:223-253.
Moriyama Y, Okamoto H, Asai H. Rubber-like elasticity and volume changes in the isolated spasmoneme of giant Zoothamnium sp. under Ca2+-induced contraction. Biophys J. 1999;76:993-1000.
Parent CA. Making all the right moves: Chemotaxis in neutrophils and Dictyostelium. Curr Opin Cell Biol. 2004;16:4-13.
Pazour GJ, Rosenbaum JL. Intraflagellar transport and cilia-dependent diseases. Trends Cell Biol. 2002;12:551-555.
Pazour GJ, Witman GB. The vertebrate primary sensory cilium is a sensory organelle. Curr Opin Cell Biol. 2003;15:105-110.
Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453-465.
Praetorius HA, Spring KR. A physiological view of the primary cilium. Annu Rev Physiol. 2003;67:515-529.
Rafelski SM, Theriot JA. Crawling toward a unified model of cell motility: Spatial and temporal regulation of actin dynamics. Annu Rev Biochem. 2004;73:209-239.
Ridge KD. Algal rhodopsins: Phototaxis receptors found at last. Curr Biol. 2002;12:R588-R590.
Ridley AJ, Schwartz MA, Burridge K, et al. Cell migration: Integrating signals from front to back. Science. 2003;302:1704-1709.
Roberts TM, Stewart M. Actin’ like actin. The dynamics of the nematode major sperm protein (msp) cytoskeleton indicate a push-pull mechanism for amoeboid cell motility. J Cell Biol. 2000;149:7-12.
Scholey JM. Intraflagellar transport. Annu Rev Cell Dev Biol. 2003;19:423-443.
Weaver AM, Young ME, Lee W-L, Cooper JA. Integration of signals to the Arp2/3 complex. Curr Opin Cell Biol. 2003;15:23-30.
Witman G. Chlamydomonas phototaxis. Trends Cell Biol. 1993;3:403-408.