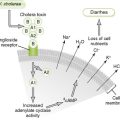
Bacterial Structure
• Their cells lack nuclei and organelles, which distinguish them from the “true” cells of eukaryotes (i.e., algae, fungi, protozoa, plants, and animals).
2. The differences between eukaryotic and prokaryotic cells, summarized in Table 6-1, are the basis for antimicrobial drugs.
TABLE 6-1
Prokaryotic Versus Eukaryotic Cells
| Characteristic | Prokaryotic Cells | Eukaryotic Cells |
| Size (approximate) (μm) | 0.5–3 | >5 |
| Cell wall | Complex structure composed of proteins, peptidoglycan, and lipids | Only in fungal and plant cells; composition differs from that of bacterial cell wall |
| Plasma (cytoplasmic) membrane | Contains no sterols (except in Mycoplasma species) | Contains sterols |
| Nuclear membrane | Absent | Present |
| Genome | Single, circular DNA molecule in nucleoid | Multiple, linear DNA molecules in nucleus |
| Organelles* | Absent | Present |
| Ribosomes | 70S (50S + 30S subunits) | 80S (60S + 40S subunits) |
| Cell division | Via binary fission | Via mitosis and meiosis |
*Include mitochondria, Golgi complex, and endoplasmic reticulum.
1. Bacterial cells range from 0.2 μm (e.g., Mycoplasma species) to 3 μm (e.g., Bacillus anthracis) in diameter.
2. The ubiquitous Escherichia coli is about 1 μm in diameter. By comparison, erythrocytes are 8 μm in diameter.
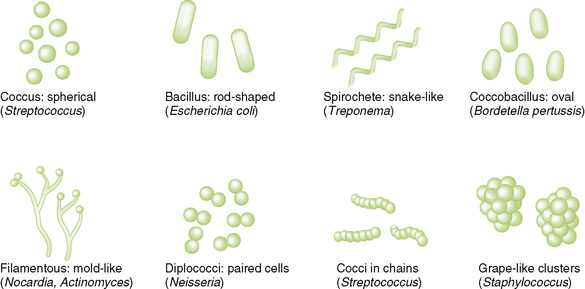
C Shape and arrangement of common bacteria (Fig. 6-1)
1. Distinguishes two major classes of bacteria (see Chapter 8)
2. Bacteria that have been starved or treated with antibiotics exhibit variable staining.
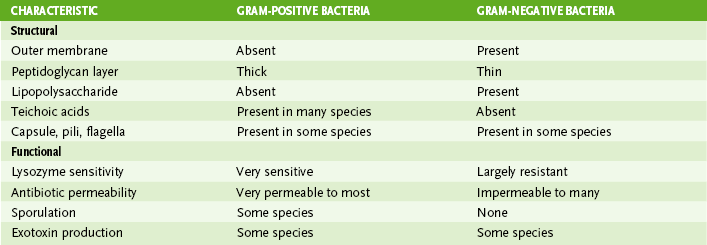
3. Gram-positive bacteria have a thick cell wall and stain purple.
4. Gram-negative bacteria have a thin cell wall and stain red.
5. Gram-resistant bacteria (e.g., Mycobacterium and Mycoplasma species) stain poorly or not at all with Gram stain.
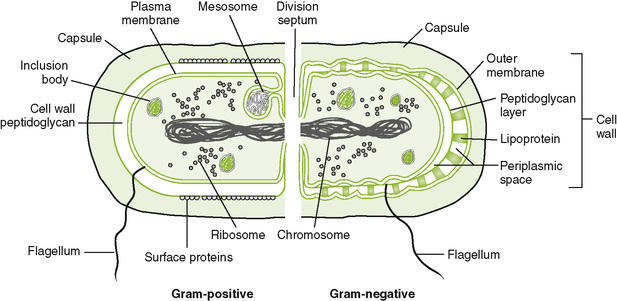
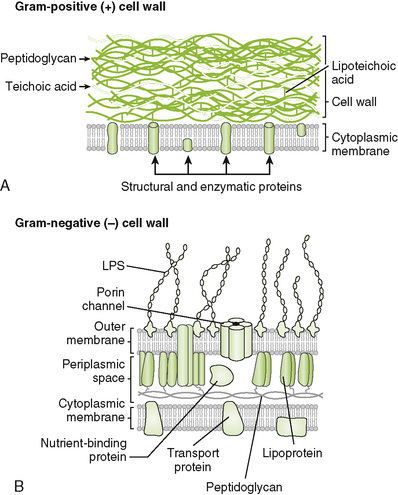
• Gram-positive and gram-negative bacteria have similar internal structures but structurally dissimilar cell envelopes (Fig. 6-2; Table 6-2).
A Internal bacterial structures
1. Nucleoid is the central region of bacterium that contains DNA.
2. Bacterial cells contain a single chromosome composed of a circular DNA molecule.
3. Because bacteria lack a nuclear membrane, transcription and translation are coupled (i.e., ribosome-mediated protein synthesis can begin while a messenger RNA [mRNA] is being produced and is still attached to the DNA).
4. Bacterial ribosomes differ in size, components, and shape from eukaryotic ribosomes and thus are a major target of antibiotic action.
5. Plasmids, which are small, circular fragments of extrachromosomal DNA, may be present and often carry antibiotic resistance genes.
• Bacterial cell envelope = cytoplasmic membrane + cell wall
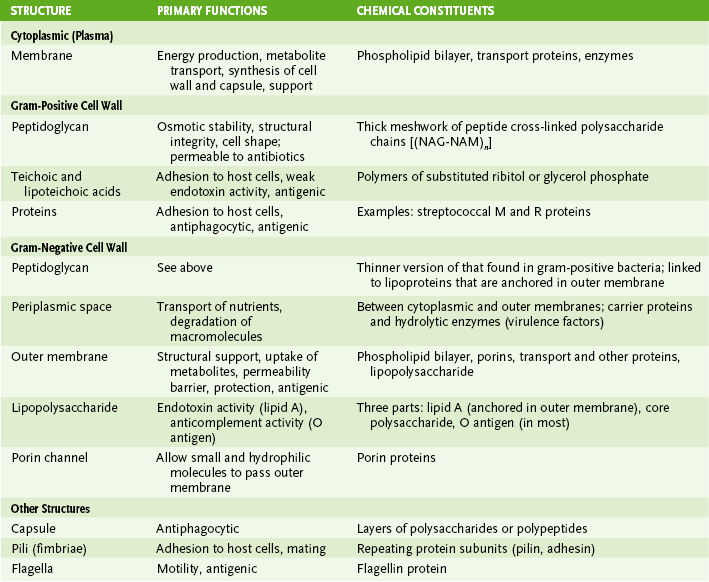
1. Cytoplasmic (cell, plasma) membrane
• Is structurally similar to eukaryotic membranes but lacks sterols, except in some mycoplasmas
• Membrane contains enzymes and other proteins that carry out energy production (e.g., electron transport chain, F1 adenosine triphosphatase), transport of nutrients (e.g., permeases), and synthesis of structural components.
2. Cell wall of gram-positive bacteria (Fig. 6-3A)
• Thick peptidoglycan layer forms a mesh-like exoskeleton essential for bacterial structure.
• Teichoic and lipoteichoic acids associated with peptidoglycan are antigenic (strain differences) and may promote adhesion to host tissue.
3. Cell wall of gram-negative bacteria (Fig. 6-3B)
• Thin peptidoglycan layer is adjacent to cytoplasmic membrane.
• Outer membrane maintains bacterial structure, acts as a permeability barrier, and provides protection against adverse environmental conditions (e.g., the digestive system of hosts).
a. Anchors LPS to bacterial surface
b. Is disrupted by polymyxin antibiotics or EDTA, which removes stabilizing Mg2+ and Ca2+ ions
• Periplasmic space, located between the outer membrane and the cytoplasmic membrane, contains degradative enzymes (e.g., β-lactamase) and nutrient-binding and transport proteins.
• Lipoproteins covalently linked to the peptidoglycan layer are inserted into the outer membrane, connecting the two structures.
• Porin channels made up of porin proteins allow small, hydrophilic molecules (including some antimicrobials) to pass through the outer membrane and limit entry of large or hydrophobic molecules.
4. Cell wall of Mycobacterium species
• Structure of peptidoglycan in mycobacteria differs slightly from that in other bacteria.
• Wax-like lipid coat containing mycolic acid surrounds the peptidoglycan-like layer.
a. This coat is responsible for virulence and antiphagocytic activity of mycobacteria.
b. Corynebacterium and Nocardia species also produce mycolic acid.
• Mycobacteria and other mycolic acid–producing bacteria can be identified with acid-fast stains.
C Other external structures (see Fig. 6-2; Table 6-3)
• Short, hair-like appendages composed of protein subunits (pilins) and anchored in plasma membrane of some bacteria
• Long, rope-like appendages that are polymers of the protein flagellin and are anchored to basal bodies in the plasma membrane of some bacteria
• Confer motility on bacteria and propel them toward food or other chemoattractants (chemotaxis)
• Express antigenic determinants that distinguish strains of organisms
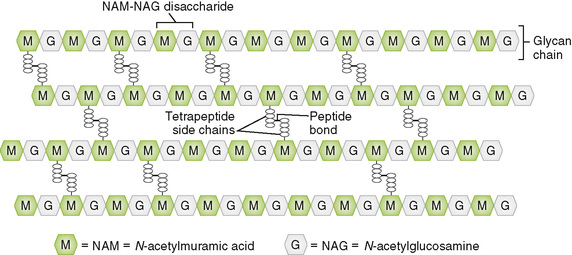
• Peptidoglycan, a rigid mesh-like polymer, is responsible for the structural integrity and shape of the bacterial cell.
A Structural parts of peptidoglycan (mucopeptide, murein)
• The basic structure consists of polysaccharide (glycan) chains with tetrapeptide or longer side chains that are cross-linked through peptide bonds (Fig. 6-4).
1. Glycan chains are linear polymers of a repeating disaccharide composed of N– acetylglucosamine (NAG) and N-acetylmuramic acid (NAM).
2. Tetrapeptide side chains contain both l amino acids and d amino acids; the latter are unusual in biologic systems.
• In a given species, all peptide side chains are identical, but their sequences may vary among species.
3. Peptide bond between a tetrapeptide attached to one glycan chain and that on another chain cross-links the two chains.
B Biosynthesis of peptidoglycan
1. Peptidoglycan is constantly being synthesized and degraded.
2. Synthesis involves four basic events.
• Assembly of NAM and NAG precursors and addition of peptide side chain to NAM in the cytoplasm
• Formation of NAG-NAM disaccharide on inner surface of the cytoplasmic membrane with the aid of bactoprenol, a long carrier molecule embedded in the inner leaflet of the membrane
• Transfer of NAG-NAM unit to growing glycan chain extends the chain and frees bactoprenol carrier for reuse
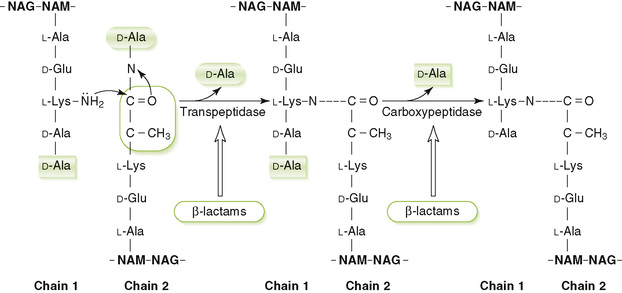
• Cross-linking of peptide side chains on adjacent glycan chains is catalyzed by transpeptidase (penicillin-binding protein [PBP]) enzymes associated with the outer surface of the membrane (Fig. 6-5)
C Antibiotics that inhibit peptidoglycan synthesis
• Degradation of peptidoglycan continues even when synthesis is inhibited, leading to cell lysis (death).
1. Vancomycin and teicoplanin are glycopeptide antibiotics that inhibit elongation of peptidoglycan chains.
2. β-Lactam antibiotics (e.g., penicillins and cephalosporins) inhibit cross-linking reaction.
3. Bacitracin interferes with recycling of bactoprenol carrier.
• LPS is the major component of the outer membrane of gram-negative bacteria and is shed into the culture medium or host tissues.
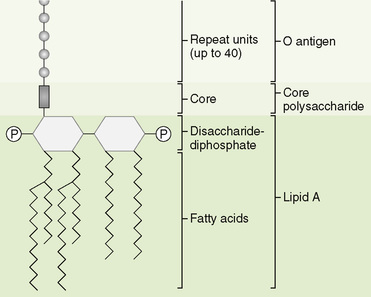
A Structural parts of LPS (Fig. 6-6)
1. Lipid A, a phosphorylated, fatty acid–modified disaccharide, is anchored in the outer membrane by its fatty acid portion.
2. Core polysaccharide, located adjacent to the outer membrane, is a branched polysaccharide of 9 to 12 sugars attached to lipid A.
3. O antigen, the outermost part of LPS, is a long, linear polysaccharide composed of 50 to 100 repeating sugar units.
1. The lipid A and core portions are enzymatically synthesized on the inner surface of the cytoplasmic membrane and then are joined together.
2. Each O antigen repeat unit is assembled on a bactoprenol molecule and then is transferred to a growing O antigen chain (similar to peptidoglycan synthesis).
3. The completed O antigen is transferred to the core lipid A structure, and the entire LPS molecule is translocated to the outer surface of the outer membrane.