Bacterial Growth, Genetics, and Virulence
I Proliferation of Bacterial Cells
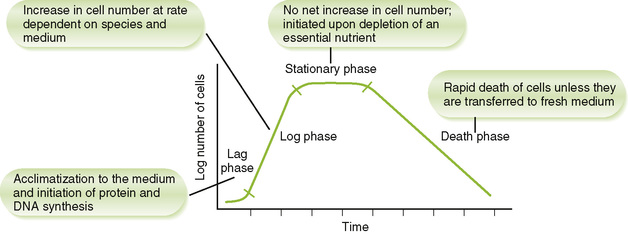
• When placed in a new medium, bacteria exhibit a typical pattern of growth and multiplication marked by four phases (Fig. 7-1).
• Different bacteria require different nutrients for growth in the body and in culture.
• Characteristic nutrient requirements and metabolic products can be used to identify bacteria.
• Bacteria divide by binary fission, a simpler process than mitotic division of eukaryotic cells.
1. Chromosome duplication is initiated at a specific sequence (replication origin) in the DNA.
• If nutrients are available, synthesis of new chromosome begins before previous chromosome synthesis is completed. Once duplication is initiated, the process is completed even when culture conditions (e.g., starvation) are detrimental to the cell.
2. Synthesis of new membrane and cell wall in the center of the cell forms a septum that eventually divides the cytoplasm into two daughter cells, each containing a complete chromosome.
D Bacterial spores (endospores)
• Spores, formed by some gram-positive bacteria, represent a dormant state that is resistant to heat, drying, and chemicals.
1. Spore formation, a variant type of cell division, is induced by depletion of essential nutrients needed for normal growth.
• Antibiotics and toxins are often produced during sporogenesis, before release of the spore from the bacterial cell.
2. Germination of spores into vegetative cells is initiated by damage to the spore coat by trauma, water, or aging and requires specific nutrients.
• Important definitions are given in Table 7-1.
TABLE 7-1
Bacterial Genetics Terminology
| Term | Definition |
| Allele | A particular example of a gene. Each variant of a given gene is a different allele of that gene. Genes that are represented by multiple alleles in a population are said to be polymorphic. |
| Cistron | Region of DNA that codes for a single protein; a complementation unit |
| Operator | Nucleotide sequence, located between the promoter and first structural gene of an operon, that binds a repressor protein |
| Operon | Bacterial transcription unit comprising a promoter, operator, and one or more structural genes |
| Plasmid | Small extrachromosomal DNA molecule capable of autonomous replication in bacteria |
| Promoter | Nucleotide sequence in an operon that is recognized by RNA polymerase |
| Replicon | Replication unit, consisting of a replication origin, a replication terminus, and the intervening coding sequence |
1. Single, double-stranded, circular molecule of DNA, containing about 5 million base pairs (or 5000 kilobase pairs)
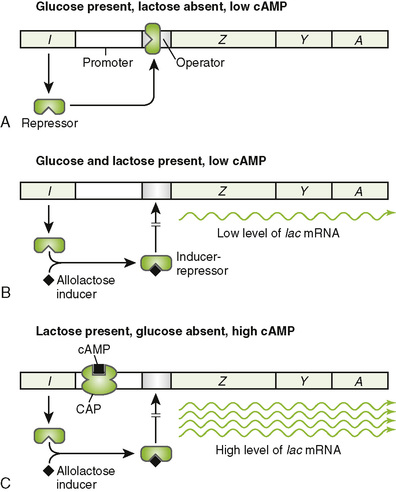
2. Operons provide coordinated control of protein-coding (structural) genes. A bacterial chromosome contains many operons.
• The enzymes in many bacterial metabolic pathways are encoded by polycistronic operons, which contain multiple structural genes.
• All the genes in a polycistronic operon are transcribed as a unit, producing a single messenger RNA (mRNA) that is translated into multiple proteins.
• Transcription of the lac operon and many other operons is controlled by presence or absence of metabolites to meet the needs of the cell (Fig. 7-2).
• DNA sequences (“jumping genes”) that can move from one position to another in a bacterial chromosome or between different molecules of DNA. These elements lack a replication origin.
C Mechanisms of genetic transfer
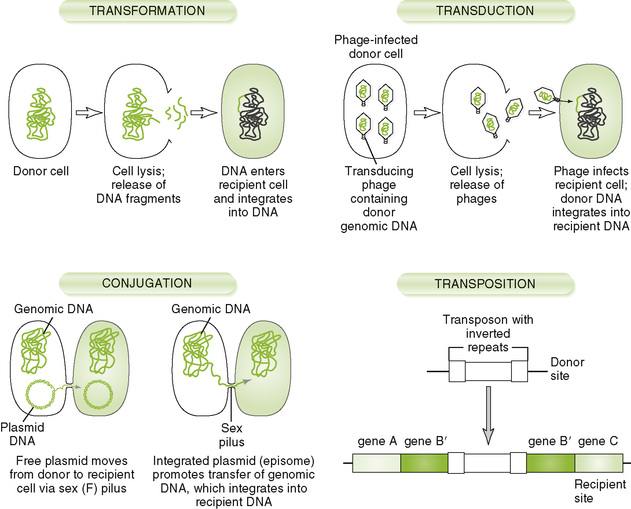
• Although bacteria replicate by binary fission, an asexual process, they have several mechanisms for transferring genetic information, as illustrated in Figure 7-3.
III Mechanisms of Bacterial Virulence (Box 7-1)
• Common routes of bacterial entry are listed in Table 7-2.
TABLE 7-2
Bacterial Entry into the Body*
| Entry Route | Examples |
| Ingestion | Bacillus cereus |
| Brucella species | |
| Campylobacter species | |
| Clostridium botulinum | |
| Escherichia coli (enterotoxigenic) | |
| Listeria species | |
| Salmonella species | |
| Shigella species | |
| Vibrio species | |
| Inhalation | Bordetella species |
| Chlamydia pneumoniae | |
| Chlamydia psittaci | |
| Legionella species | |
| Mycobacterium species | |
| Mycoplasma pneumoniae | |
| Nocardia species | |
| Trauma/needlestick | Clostridium tetani |
| Pseudomonas species | |
| Staphylococcus aureus | |
| Arthropod or animal bite | Borrelia species |
| Coxiella species | |
| Ehrlichia species | |
| Francisella species | |
| Rickettsia species | |
| Yersinia pestis | |
| Sexual transmission | Chlamydia trachomatis |
| Neisseria gonorrhoeae | |
| Treponema pallidum | |
| Transplacental | Treponema pallidum |
• Disease results from tissue destruction, compromised organ function, or host defense responses that produce systemic symptoms (e.g., fever, nasal congestion, headache, lethargy, and loss of appetite).
1. Common pili (fimbriae) are the major structures involved in adherence of enteric bacteria and N. gonorrhoeae.
• Bacteria that lack pili may possess other cell surface adhesins (e.g., lectin protein, lipoteichoic acid, and slime).
2. Adhesin molecules, whether present on pili or elsewhere, interact with receptors on host cells.
• Refers to bacteria breaking through tissue barriers and colonizing tissues (Box 7-2)
1. Some enteric bacteria have mechanisms for invading various portions of the gastrointestinal tract.
2. Invasion of normally sterile sites even by normal microbial flora can cause diseases such as bacteremia, meningitis, encephalitis, and pneumonia.
3. Colonization of a tissue can cause its destruction or dysfunction or can be a source of spread in the body or release of toxins, including endotoxins.
• Caused in part by bacterial products.
1. Tissue-damaging metabolites (e.g., acids, gases, and other byproducts) may be formed during bacterial growth.
2. Degradative enzymes and cytolytic exotoxins are released by many bacteria.
• Clostridium perfringens: lecithinase (α toxin)
• Streptococci: hemolysin, streptokinase, hyaluronidase
• Staphylococci: hyaluronidase, fibrinolysin, lipases, enterotoxins
3. Exotoxins that disrupt normal cellular metabolism (see section G).
4. Intracellular growth can disrupt cell function (e.g., mycobacteria)
5. Bacterial growth on top of tissue can damage and disrupt tissue function (e.g., coagulase-negative staphylococci or viridans streptococci subacute endocarditis)
D Circulation through the blood (bacteremia)
1. Primary mechanism for spreading bacteria throughout the body
2. Tissue damage promotes bacterial spread, especially if blood vessels are damaged.
E Pathogen-associated molecular patterns (PAMPs)
1. PAMPs are repetitive microbial structures that bind to toll-like receptors (TLRs) and other pathogen pattern receptors on various cells, activate macrophages and dendritic cells, and promote acute phase cytokine release.
2. Examples of PAMPs: peptidoglycan, lipopolysaccharide (LPS), lipoteichoic acid, teichoic acid, flagellin, and CpG oligodeoxynucleotides
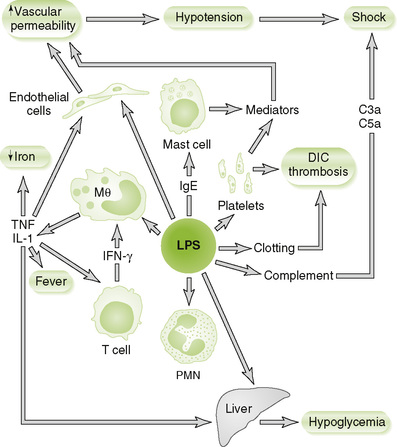
1. The lipid A component of LPS
2. Is present in the cell wall of all gram-negative bacteria
3. Is released during early stages of infection with gram-negative bacteria
4. Initiates complement and clotting pathways
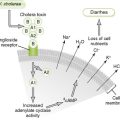
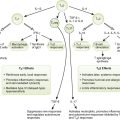
5. LPS is a strong PAMP and binds to TLRs on various cells to induce release of mediators that cause sepsis (Fig. 7-4; Box 7-3).
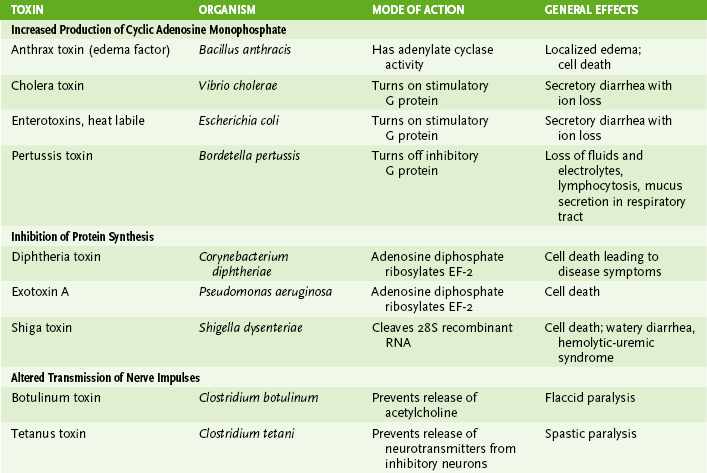
• Secreted by certain gram-positive and gram-negative bacteria (Table 7-3)
TABLE 7-3
| Property | Endotoxin | Exotoxins |
| Produced by | Gram-negative bacteria | Certain gram-positive and gram-negative bacteria |
| Location of genes | Bacterial chromosome | Bacterial chromosome, plasmid, bacteriophage |
| Composition | Lipid A part of lipopolysaccharide in cell wall | Polypeptide (secreted) |
| Stability | Heat stable | Heat labile (usually) |
| Major biologic actions | Induces tumor necrosis factor-α, interleukin (IL-1), IL-6; initiates complement and clotting pathways | Inhibit protein synthesis, block neuro-transmitter release, increase cyclic adenosine monophosphate level, etc. |
| Clinical effects | Fever, hypotension, shock, disseminated intravascular coagulation | Various, depending on type (see Table 7-4) |
| Used as vaccine | No: poorly antigenic | Yes (inactivated toxoids): highly antigenic |
1. Cytolytic exotoxins are tissue-degrading enzymes.
2. A-B toxins are composed of one or more B subunits, which bind to the cell surface, and an A subunit, which enters the cell and acts on it (Table 7-4).
3. Superantigen exotoxins activate T cells in the absence of antigen by cross-linking the T cell receptor and class II major histocompatibility complex on antigen-presenting cells.
• Cause inappropriate release of cytokines (e.g., interleukin-1 [IL-1], IL-6, tumor necrosis factor-α, and other cytokines), with possible life-threatening autoimmune responses and death of T cells
• Toxic shock syndrome toxin (TSST) of S. aureus, staphylococcal enterotoxins, and erythrogenic toxin of Streptococcus pyogenes are prime examples.
1. It can spread through a bacterial population by transfer of antibiotic-resistance genes (see Fig. 7-3), or antibiotic may eliminate competing, nonresistant organisms.
I Escape of host attempts at elimination
• Evasion of host defenses permits bacteria to remain in the host longer, thereby causing more damage.
1. Encapsulation with polysaccharide (slime) layer (Box 7-4)
• Capsules are poorly antigenic, deter phagocytosis because they are slippery and tear away, and protect against degradation in phagolysosomes of macrophages and neutrophils.
• Virulence of some encapsulated bacteria (e.g., S. pneumoniae and N. meningitidis) is lost in mutants lacking a capsule.
2. Biofilms are sticky webs of polysaccharide that can protect the bacteria from host defenses and antimicrobials.
• Sufficient bacteria must be present to make a biofilm (quorum sensing).
• P. aeruginosa and other bacteria make biofilms. Dental plaque is a biofilm.
3. Intracellular growth without inactivation (especially in macrophages)
• Organisms that can grow and multiply within host cells escape immune detection.
a. Inhibition of phagolysosome fusion (M. tuberculosis), protective outer layers (M. tuberculosis), and resistance to lysosomal enzymes allow replication within cells.
b. Obligate intracellular bacteria: Rickettsia, Chlamydia, Coxiella species
• Chronic stimulation of immune response may result in granuloma formation
4. Reduction of phagocytic cell function
• Inhibition of phagocytosis by antiphagocytic capsule and certain cell surface proteins (e.g., M protein of S. pyogenes)
• Reduction in phagolysosomal killing of phagocytosed bacteria by capsules, inhibition of phagosome-lysosome fusion (M. tuberculosis), and catalase detoxification of peroxide (S. aureus)
• Killing of phagocytic cells by exotoxins (e.g., streptolysins of S. pyogenes, lethal toxin of anthrax, and α toxin of C. perfringens)
• Immunoglobulin A (IgA)-degrading proteases produced by some bacteria permit them to colonize mucosal surfaces: N. gonorrhoeae, N. meningitidis, S. pneumoniae, and Haemophilus influenzae.
• IgG-binding surface proteins (e.g., protein A of S. aureus) protect bacterial cells from antibody action.
6. Inhibition of complement action
• Antigen in LPS of gram-negative bacteria may protect cells from complement-mediated lysis by preventing access to the membrane.
7. Antigenic variation by DNA rearrangement involving homologous recombination
IV Antibacterial Immunopathogenesis
• Host immune responses cause the disease for some bacteria.
1. Complement, neutrophils, macrophages, and other responses to bacteria lead to inflammation.
2. Cell-mediated immune responses induced to intracellular infections (M. tuberculosis) may lead to granulomas.
B Bacteria that induce tissue-damaging immune responses
• More common for viruses than for bacteria
1. Chlamydia species (lymphogranuloma venereum)
C Cross-reacting antibacterial antibodies
• Rheumatic fever, a sequela to streptococcal infections, is caused by antibodies to M protein that cross-react with and initiate damage to the heart.
D Deposition of immune complexes
• Poststreptococcal glomerulonephritis results from accumulation of antigen-antibody complexes in the glomeruli of the kidneys.