CHAPTER 33 Actin and Actin-Binding Proteins
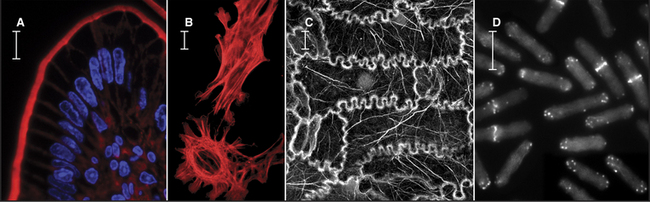
Actin filaments form a cytoskeletal and motility system in all eukaryotes (Fig. 33-1). Cross-linked actin filaments resist deformation, transmit forces, and restrict diffusion of organelles. A network of cortical actin filaments excludes organelles (Fig. 33-2C), reinforces the plasma membrane, and restricts the lateral motion of some integral membrane proteins. The cortex varies in thickness from a monolayer of actin filaments in red blood cells (see Fig. 7-10) to more than 1μm in amoeboid cells (Fig. 33-2C). Like fingers in a glove, bundles of actin filaments support slender protrusions of plasma membrane called microvilli or filopodia (Fig. 33-2B). Microvilli expand the cell surface for transport of nutrients and participate in sensory processes, including hearing. The actin cytoskeleton complements and interacts physically with cytoskeletal structures composed of microtubules (see Chapter 34) and intermediate filaments (see Chapter 35).
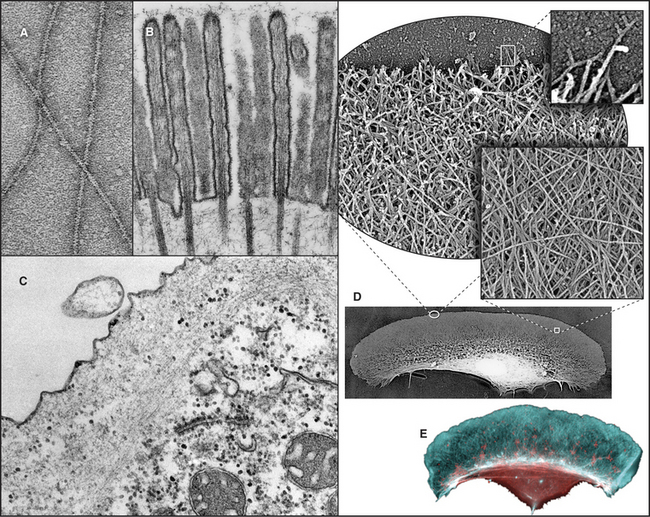
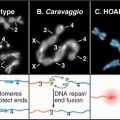
Figure 33-2 electron micrographs of actin filaments. A, Filaments of purified actin prepared by negative staining.B, A thin section of an intestinal epithelial cell illustrating finger-like microvilli with tightly packed bundles of actin filaments linked to the surrounding plasma membrane by myosin-I. The barbed ends of these filaments (see Fig. 33-9) are located at the tips of the microvilli.C, A thin section of Acanthamoeba showing the actin filament meshwork in the cortex beneath the plasma membrane.D–E, Cultured fish scale keratocytes, fixed while actively migrating toward thetop of the figure. D, Electron micrograph of a whole mount of a cell illustrating the meshwork of branched filaments near the leading edge and longer, unbranched filaments deeper in the cytoplasm. Most filaments are oriented with their barbed ends forward.E, Fluorescence micrograph with phalloidin staining actin filaments (blue) and antibodies staining myosin II (red).
(A, Courtesy of U. Aebi, University of Basel, Switzerland. B, Courtesy of L. Tilney, University of Pennsylvania, Philadelphia, and M. Mooseker, Yale University, New Haven, Connecticut. D–E, Courtesy of T. Svitkina and G. Borisy, University of Wisconsin, Madison.)
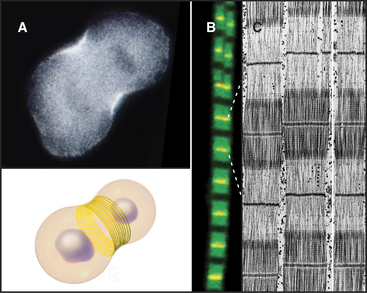
Actin contributes to cell movements in two ways. First, polymerization and depolymerization of the network of actin filaments just inside the plasma membrane contribute to the extension of pseudopods, cell locomotion (Fig. 33-2D-E), and phagocytosis (see Fig. 22-3). Second, actin filaments are tracks for movements of the myosin family of motor proteins (see Fig. 36-7). Actin filaments and myosin filaments form the highly ordered, stable contractile apparatus of muscles (Fig. 33-3B; also see Fig. 39-3), as well as the transient contractile ring that pinches the two daughter cells apart at the end of mitosis (Fig. 33-3A; also see Fig. 44-23). Myosins also power movements of membranes and other cargo along actin filaments, complementing organelle movements by other motors along microtubules (see Fig. 37-1). Actin, myosin, and accessory proteins form intracellular bundles called stress fibers (Fig. 33-1B) that apply tension between adhesive junctions on the plasma membrane (see Fig. 30-11), where cells attach to each other or to the extracellular matrix. Stress fibers are prominent in tissue culture cells grown on glass or plastic and in endothelial cells lining major arteries.
Actin and myosin are thought to be among the five most abundant eukaryotic proteins on the earth. Actin is often the most abundant protein in a cell, composing up to 15% of total protein, and the many types of actin-binding proteins may account for another 10% of cellular protein. In muscle, actin and myosin constitute more than 60% of the total protein. Given this abundance, it is curious that actin was discovered in muscle only in the 1940s and in nonmuscle cells in the late 1960s. Since the 1970s, scientists have discovered new actin-binding proteins every year, but the inventory is probably still incomplete. Genetic defects in components of the actin cytoskeletal and motility system cause many human diseases, including muscular dystrophy (see Table 39-2), hereditary fragility of red blood cells (i.e., hemolytic anemias, see Fig. 7-10), and hereditary heart diseases called cardiomyopathies (see Table 39-4).
Actin Molecule
Actin is folded into two domains that are stabilized by an adenine nucleotide lying in between (Fig. 33-4). The polypeptide of 375 residues crosses twice between the two domains, with the N- and C-termini located near each other. The two domains are folded similarly, suggesting that the actin gene arose by duplication of an ancestral gene. Remarkably, the adenine nucleotide binding site, fold, and overall shape of actin closely resemble those of two other proteins with very different functions: the glycolytic enzyme hexokinase (see Fig. 3-12) and the heat shock protein Hsc70. All three proteins might have evolved originally in prokaryotes from the same primordial nucleotide-binding protein.
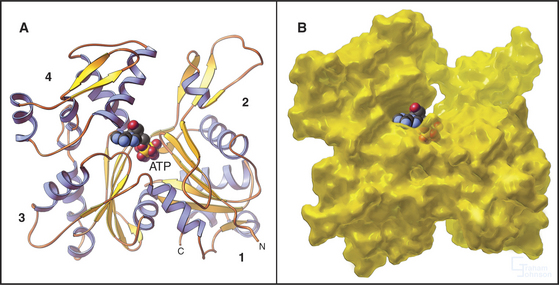
Figure 33-4 atomic structure of actin. A, Ribbon model showing the polypeptide fold and the location of Mg-ATP, shown as space-filling. Numbers indicate the four subdomains.B, Surface rendering. ATP is almost completely buried in the cleft between the two lobes of the protein, where it makes extensive contacts with the protein. The barbed end of the molecule (Fig. 33-9) is at the bottom in this orientation.
(PDB file: 1ATN. Reference: Kabsch W, Mannherz HG, Suck D, et al: Atomic structure of the actin-DNase I complex. Nature 347:37–44, 1990.)
Actin binds adenosine triphosphate (ATP) or adenosine diphosphate (ADP) and a divalent cation, Mg2+ in cells, with nanomolar affinity. The affinity of actin for ATP is higher than that for ADP, so given the higher concentration of ATP in cells, unpolymerized actin is saturated with ATP. The bound nucleotide exchanges relatively slowly with nucleotide in the medium (Fig. 33-11). Actin monomer–binding proteins can inhibit or accelerate nucleotide exchange. Bound nucleotide stabilizes the molecule but is not required for polymerization in vitro. ATP-actin and ADP-actin polymerize at different rates.
Posttranslational modifications of actins include acetylation of the N-terminus and (in most cases) methylation of histidine-68. In some insect flight muscles, the small protein ubiquitin (see Fig. 23-7) is attached covalently to about one in six actin molecules, yielding a 55-kD polypeptide that is incorporated with unmodified actin into filaments. Some invertebrate actins are phosphorylated on tyrosine-211. The functional significance of these modifications is still being investigated.
Actin genes originated in prokaryotes, where they are required for rod-shaped bacteria to maintain their asymmetric shapes. Other bacterial actins help to segregate DNA plasmids to the two daughters during cell division. Eukaryotic actin genes are highly conserved, but through divergent evolution, they encode subtly different proteins, some with novel functions. Most organisms have multiple actin genes, and all known actin isoform diversity arises from multiple genes rather than from alternative splicing of mRNAs. Humans have six actin genes; Dictyostelium has more than ten; but budding yeast has only one. Muscle actin genes diverged from cytoplasmic actins in primitive chordates (see Fig. 2-9). To fulfill special developmental functions, plant actin genes diverged among themselves more than animal actin genes.
The biochemical similarities of actin isoforms are more impressive than their differences (Fig. 33-5). The sequences of pairs of actins are generally more than 90% identical, even between highly divergent species. Humans express β and γ isoforms in nonmuscle cells and four different α and β isoforms in various muscle cells. Many nonmuscle cells express both the β and γ isoforms, but red blood cells use only β-actin.
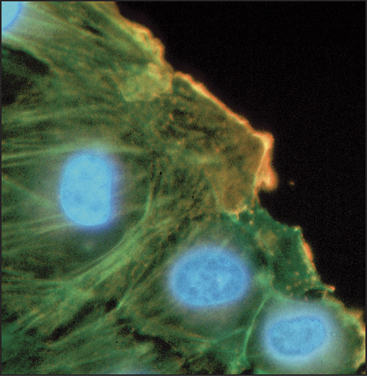
In every case that was examined, actin isoforms copolymerize in the test tube, so it is remarkable that cells can sort actin isoforms into different structures. For example, b-actin is concentrated near the plasma membrane of cultured cells, whereas g-actin is concentrated in stress fibers (Fig. 33-6). In muscle, α-actin forms the thin filaments of the contractile apparatus, whereas g-actin localizes around mitochondria. Even the full array of known actin-binding proteins cannot yet explain how cells prevent copolymerization of the isoforms or concentrate isoforms at different locations.
Actin-Related Proteins
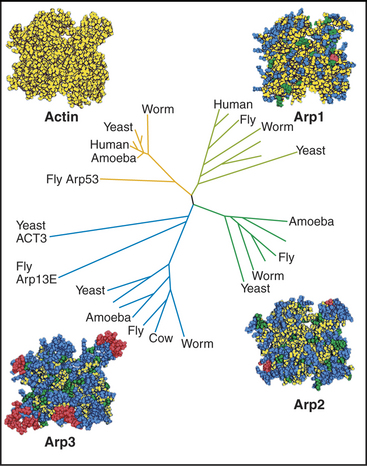
After believing for 20 years that actins are one of the most evolutionarily conserved protein families, scientists discovered several families of highly divergent actin-related proteins (Arps) in the 1990s (Fig. 33-6). Genes for Arps diverged from actin genes after the earliest branches in the eukaryotic tree and are now found in eukaryotic species ranging from amoebas to humans. Arps share with actin the fold of the polypeptide chain and residues forming the nucleotide-binding site, but fewer than 60% of the residues are identical to actin. Divergent surface residues allow Arps to participate in molecular interactions different from actin. Arp1 forms a short filament as part of the dynactin complex that promotes cargo movement by the microtubule motor dynein (see Fig. 37-2). Arp2 and Arp3 are two of seven subunits in a protein complex that nucleates branched actin filaments in the cell cortex (Fig. 33-13). Eight additional types of Arps are widespread in eukaryotes. Several participate in complexes that regulate chromatin structure.
Actin Polymerization
Actin filaments are polarized, owing to the uniform orientation of the asymmetrical subunits along the polymer (Fig. 33-7). One end is called the barbed end, the other is called the pointed end. This nomenclature arises from the asymmetrical arrowhead pattern created when myosin bind along the length of actin filaments (Fig. 33-8). The helical arrangement of subunits in actin filaments was originally revealed in the 1960s by electron microscopy and X-ray fiber diffraction of whole muscle and actin gels. These low-resolution data are used to orient the atomic structure of the actin monomer in current models.
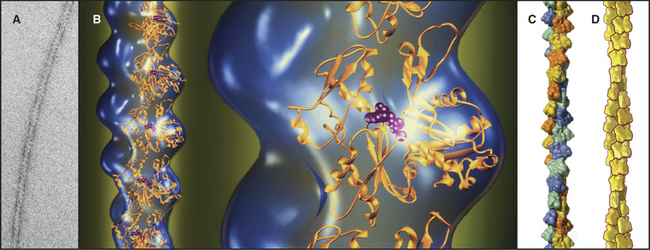
Figure 33-7 structure of the actin filament. A, Electron micrograph of a negatively stained actin filament. B, Reconstruction of an actin filament by image processing of electron micrographs (blue) with ribbon models of the subunits(gold) along one strand of the double helix. One subunit is enlarged to the right. The pointed end of the subunits with the nucleotide cleft is at thetop, and the faster-growing barbed end is at thebottom. This orientation of the actin molecule in the filament uniquely accounts for the X-ray fiber diffraction pattern of oriented filaments and agrees with electron microscopy with probes on specific actin residues and with chemical cross-linking between residues of adjacent subunits. C, Surface rendering of the molecular model. Subunits in the two long-pitch helices are shown asyellow-orange andblue-green (see Fig. 5-5 for nomenclature). The short pitch helix, including every subunit, follows a yellow-green-orange-blue pattern.D, Scale drawing used throughout this text.
(A–B, Courtesy of U. Aebi, University of Basel, Switzerland. C, Courtesy of R. Milligan, Scripps Research Institute, La Jolla, California.)
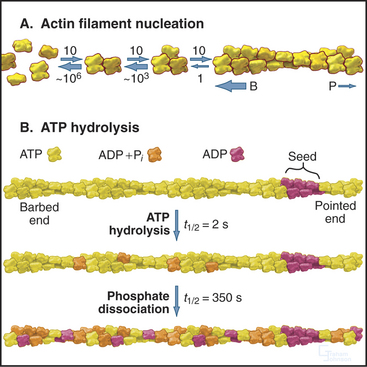
Actin self-assembles into filaments by means of a series of bimolecular reactions (Fig. 33-9; see also Fig. 5-6). Actin is isolated from cells as a monomer at low salt concentrations. Physiological concentrations of monovalent and divalent cations bind to low-affinity sites on actin and promote polymerization. In vitro, actin trimers appear to be the nucleus that initiates polymer growth in the sense that the reactions that are required to form trimers are very unfavorable in comparison with reactions for elongation of polymers larger than trimers. To initiate new filaments, cells use regulatory proteins to overcome these unfavorable nucleation reactions.
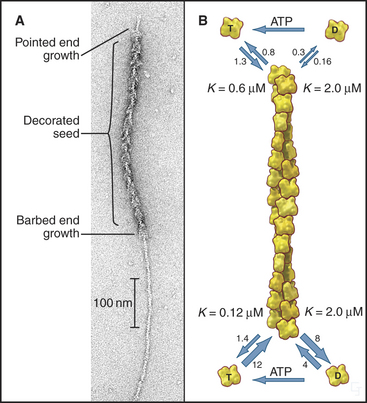
Actin filaments grow and shrink by the addition and loss of subunits at the two ends of the polymer. The reactions at the two ends have different rate constants (Fig. 33-8). Association of subunits is rapid at both ends. Subunit association is a diffusion-limited reaction (see Chapter 4) at the rapidly growing barbed end and somewhat slower at the other end. Subunit dissociation is relatively slow at both ends, between 0.3 and 8 subunits per second. The rates of these reactions depend on the nucleotide bound to the monomer associating with or dissociating from a filament.
In the presence of ATP, purified actin assembles almost completely, leaving as monomers the critical concentration of about 0.1μM ATP-actin. The critical concentration is the monomer concentration giving equal rates of association and dissociation, 1.4s−1 at the barbed end (see Figure 5-6). The critical concentration for ADP-actin is about 20 times higher than for ATP-actin.
Hydrolysis of bound ATP and dissociation of the g-phosphate during assembly modifies the behavior of actin filaments, including their affinity for regulatory proteins. Following incorporation of an ATP-actin subunit into a filament, bound ATP is hydrolyzed irreversibly to ADP and phosphate with a half time of 2s (Fig. 33-9). These ADP-Pi subunits behave much like ATP subunits. Phosphate dissociates slowly and reversibly over several minutes. This yields filaments with a core of subunits with tightly bound ADP. At the millimolar concentrations of phosphate in cytoplasm, phosphate rebinds to some ADP-actin subunits.
The critical concentrations for ATP-actin differ at the two ends of the filament. This results from differences in the probability of nucleotide hydrolysis and phosphate release at the two ends, which is more likely to expose ADP-subunits at the pointed end. At steady state in the presence of ATP, the actin monomer concentration falls between the critical concentrations at the two ends. Though the polymer and monomer concentrations remain constant, net addition of subunits at the barbed end and net loss of subunits at the pointed end result in the slow migration of subunits through the polymer from the barbed end to the pointed end. This process is called treadmilling. Neither end exhibits rapid fluctuations in length like those of microtubules (see Fig. 34-7).
In many cell types, the barbed ends of the filaments are associated with the plasma membrane (Fig. 33-2). Actin filaments in muscle are also anchored at their barbed ends (Fig. 33-3C; also see Fig. 39-3). This makes mechanical sense, as actin filaments sustain tension better than compression and because (with one interesting exception) all known myosins pull filaments in a direction away from the barbed end. The plasma membrane is a prominent actin filament–anchoring site, allowing force that is produced in the cytoplasmic actin network to change the shape of the membrane and to be transmitted to the substrate on which the cell sits or to adjacent cells (see Fig. 38-5).
Actin-Binding Proteins
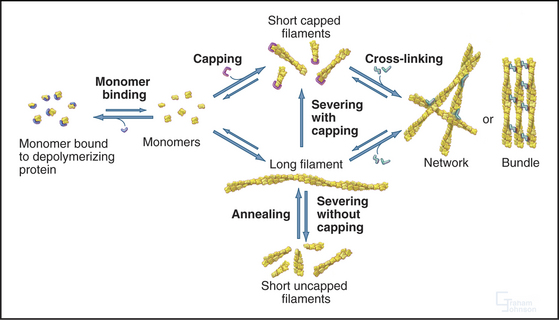
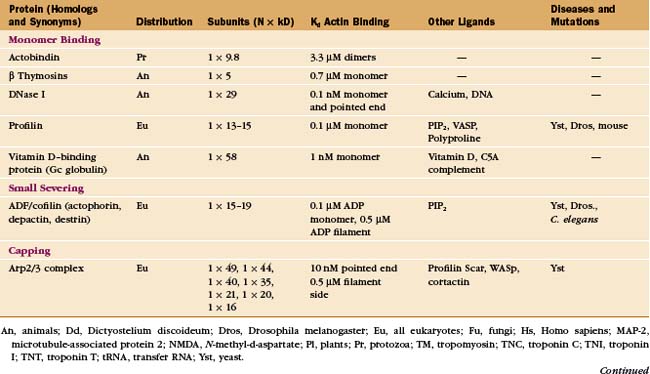
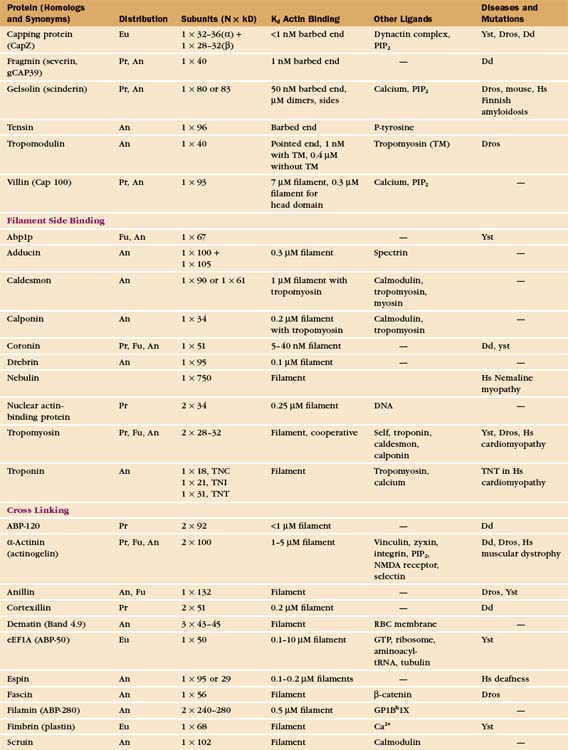
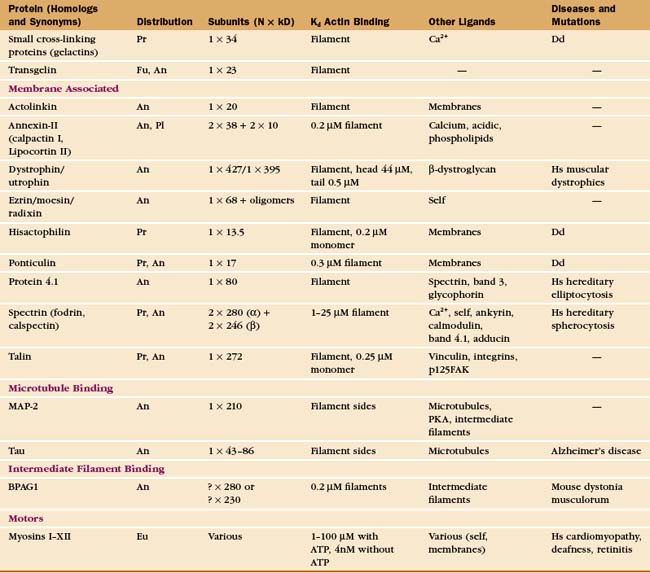
In contrast to the slow treadmilling of filaments of purified actin, actin filaments in live cells can polymerize and depolymerize rapidly under the control of more than 60 families of actin-binding proteins (Fig. 33-10 and Appendix 33-1). Broadly, these proteins fall into families that bind monomers, sever filaments, cap filament ends, nucleate filaments, cross-link filaments, stabilize filaments, or move along filaments. Like actin, actin-binding proteins are ancient. Many families arose in early eukaryotes and are found in protozoa, yeast, plants, and vertebrates.
Actin Monomer–Binding Proteins
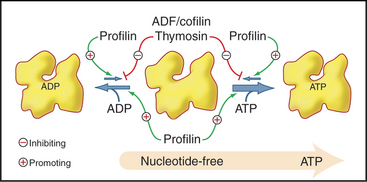
Profilins are abundant proteins, found in all branches of the eukaryotic tree. Cells require three different profilin activities for viability: binding actin monomers, catalyzing the exchange of nucleotides bound to actin (Fig. 33-11) and binding to polyproline sequences on other protein such as formins (Fig. 33-12). Profilins also bind acidic membrane lipids (polyphosphoinositides). The nucleotide bound to actin monomers determines the affinity for profilin: highest for nucleotide-free actin monomers, followed by ATP-actin and ADP-actin. Profilins bind to the barbed end of actin monomers, thereby sterically blocking nucleation and pointed end elongation but not association of the profilin-actin complex with the barbed end of filaments. Profilin dissociates rapidly after the profilin-actin complex binds to a barbed end.
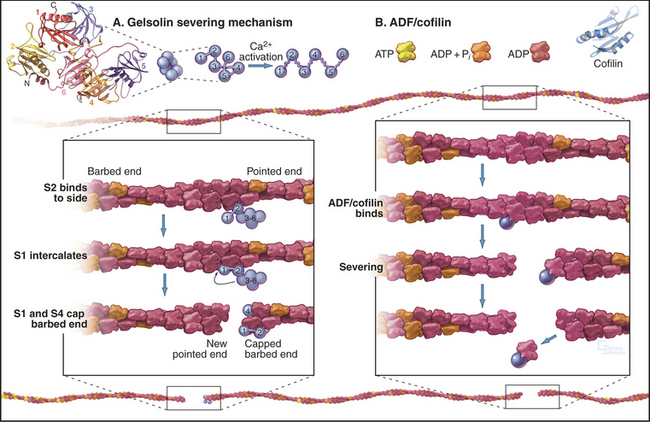
Members of the ADF/cofilin family are essential for the viability of many eukaryotes. They bind ADP-actin monomers with higher affinity than ATP-actin and inhibit nucleotide exchange but not polymerization. Their major roles are to bind and destabilize ADP-actin filaments (Figs. 33-15 and 33-19) and to nucleate filaments.
Actin Filament Nucleation Factors
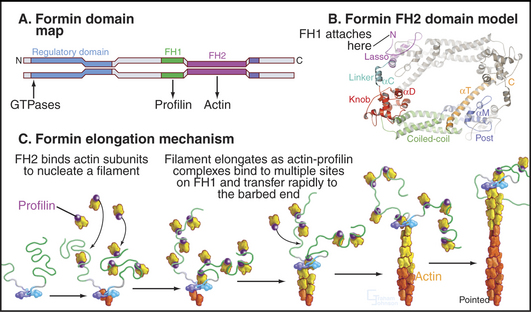
Formins initiate unbranched actin filaments that are incorporated into the contractile ring and bundles of actin filaments such as cables in yeast (see Fig. 37-11) and stress fibers in animal cells (Fig. 33-1B). Each of the three formins in fission yeast assembles actin filaments for a specific function: the cytokinetic contractile ring, interphase actin cables, or mating structures. Thus, the 15 different mammalian formins are also likely to have specific functions. These proteins have in common a formin homology-2 (FH2) domain (Fig. 33-12). Pairs of FH2 domains form a doughnut-shaped ring that fits around the barbed end of an actin filament. Interactions with two actin monomers nucleate a filament that grows by adding subunits to the barbed end. An FH2 domain can track faithfully on the growing end as it adds thousands of new subunits. When associated with a membrane in a cell or a microscope slide in an experiment, a formin molecule can extrude a growing actin filament for many minutes. Profilin enhances elongation by targeting profilin-actin complexes to multiple polyproline sequences in the FH1 domain adjacent to FH2. Some well-studied formins are autoinhibited by interactions between parts of the polypeptide flanking the FH1FH2 domains. Rho family GTPases regulate foramins by overcoming this autoinhibition.
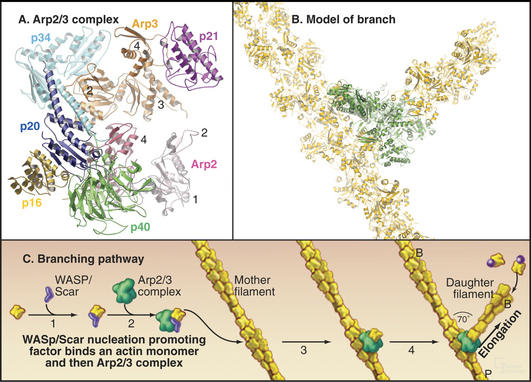
Arp2/3 complex consists of two actin-related proteins (Arp2 and Arp3) tightly bound to five novel proteins (Fig. 33-13). Arp2/3 complex binds to the side of actin filaments and forms branches by nucleating filaments that grow at their free barbed ends. The complex caps the pointed end of the new filament and attaches it to the side of the older filament. Growth of the free barbed ends of these branches produces the force that pushes the plasma membrane forward at the leading edge of motile cells (Fig. 33-2D).
Spire was discovered in Drosophila as a gene required for the development of eggs and embryos and later found in other metazoans but not fungi or protozoa. Spire proteins have multiple domains (corresponding to the V domains in Fig. 33-17A) that bind actin monomers and stabilize oligomers on the pathway to nucleation of unbranched filaments. The biological functions of these proteins are being investigated.
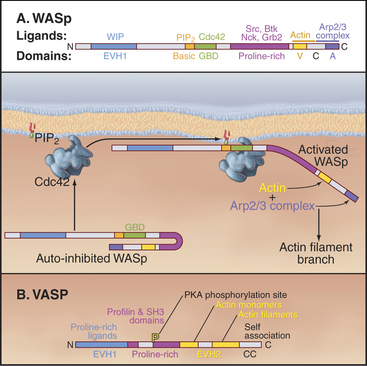
Figure 33-17 adapter proteins. A, WASp (Wiskott-Aldrich syndrome protein). Domains called V (for verprolin homology), C (for connecting), and A (for acidic) activate Arp2/3 complex. VC binds an actin monomer, and CA binds Arp2/3 complex. C also binds intramolecularly to the GBD (GTPase-binding domain), autoinhibiting VCA. Membrane-bound Rho-family GTPases and polyphosphoinositides compete C from GBD, releasing VCA to interact with actin and Arp2/3 complex. SH3 domain proteins such as Nck also activate WASp by binding the proline-rich domain. The N-terminal EVH1 (Enα-VASP homology) domain binds WIP (WASp interacting protein), the mammalian homolog of yeast verprolin.B, VASP (vasodilator-stimulated phosphoprotein). The EVH1 domain (see Fig. 25-11B) binds polyproline ligands, including vinculin and zyxin in focal contacts; the proline-rich domain binds profilin, profilin-actin complexes, and proteins with certain SH3 domains; the EVH2 domain binds both actin monomers and filaments; and the C-terminal coiled-coil (CC) mediates the formation of VASP tetramers.
Actin Filament–Capping Proteins
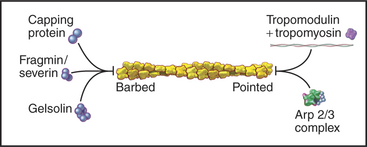
Capping proteins bind to either the barbed or pointed end of actin filaments, where they block subunit addition and dissociation (Fig. 33-14). Some capping proteins also stimulate the formation of new filaments and/or sever actin filaments (Fig. 33-15).
Gelsolins consist of six domains with similar folds but different sequences and functions. They bind tightly to the sides and barbed ends of actin filaments, blocking both the dissociation and association of actin subunits. Gelsolin also binds actin dimers, forming a nucleus that grows at the pointed end. Phosphatidyl 4,5-bisphosphate (PIP2) competes with actin for binding gelsolin. Three-domain capping proteins, such as fragmin and severin, are similar in structure and function to the first three domains of gelsolin. Genes for these capping proteins, found widely across the phylogenetic tree, are likely to have duplicated during evolution to give rise to gelsolin genes.
Heterodimeric capping proteins consist of two subunits of approximately 30 kD. They cap barbed ends with high affinity, independent of Ca2+, and promote nucleation of new pointed ends by stabilizing small actin oligomers. As with gelsolins, PIP2 inhibits capping by these proteins. Heterodimeric capping proteins are found in most eukaryotic cells. In striated muscle, they cap the barbed end of actin filaments in the Z disk (see Fig. 39-5).
Tropomodulin caps the pointed end of stable actin filaments in muscle, red blood cells, and other cells of higher organisms. High-affinity binding to pointed ends requires tropomyosin, an α-helical protein that binds along the length of actin filaments (see Fig. 39-4).
Actin Filament–Severing Proteins
Three classes of proteins just introduced—gelsolin, fragmin/severin, and ADF/cofilin—also sever actin filaments into short fragments (Fig. 33-15). Domain 2 of gelsolin binds to the side of an actin filament, positioning domain 1 to bind between subunits and disrupt the filament. This requires micromolar concentrations of Ca2+. One gelsolin isoform is found inside cells; another is secreted into blood plasma, where it may sever actin filaments released from damaged cells. Fragmin and severin from slime molds (but not their vertebrate homologs) have similar Ca2+-dependent severing activity. Independent of Ca2+, ADF/cofilins bind ADP-actin subunits in filaments and promote severing and depolymerization.
Proteins That Bind the Side of Actin Filaments
Tropomyosin, nebulin, and caldesmon are extended proteins that bind along the sides of actin filaments. Tropomyosin increases the tensile strength of actin filaments; in striated muscles, it is also an essential component of the Ca2+-sensitive regulatory machinery that controls the interaction of myosin and actin (see Fig. 39-4). Nebulin may determine the length of the actin filaments in skeletal muscle (see Chapter 39). Caldesmon, together with tropomyosin and Ca2+-calmodulin, regulates interaction of actin and myosin in smooth muscle (see Fig. 39-21) and nonmuscle cells. Phosphorylation of caldesmon by cell cycle kinases and may play a role in the reorganization of actin filaments during mitosis.
Actin Filament Cross-Linking Proteins
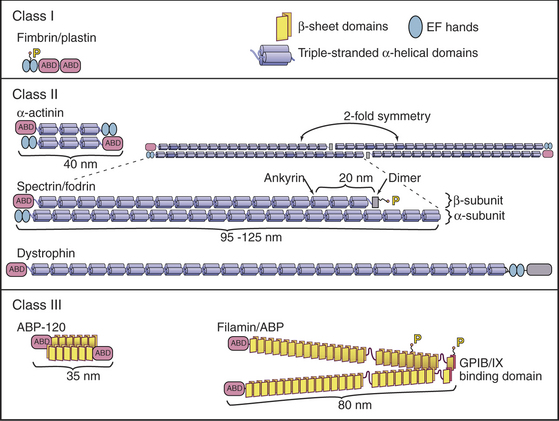
Possession of two actin-binding sites enables cross-linking proteins (Fig. 33-16) to bridge filaments and to stabilize higher-order assemblies of actin filaments. Some have a greater tendency to cross-link filaments in regular bundles, like those in microvilli (Fig. 33-2A), but depending on protein concentrations and filament lengths, most of these proteins can promote the formation of both random networks and regular bundles of filaments.
Many of these proteins share a homologous actin binding domain associated with other domains that form dimers. α-Actinin is found in the cortical actin network, at intervals along stress fibers, on the cytoplasmic side of cell adhesion plaques (see Fig. 30-11), and in the Z-disk of striated muscles (see Fig. 39-5). Fimbrin and villin (a relative of gelsolin with an extra actin-binding site) stabilize the regular actin filament bundles in microvilli. Filamin cross-links filaments in the cortex of many cells and also anchors these filaments to an integrin, a plasma membrane receptor for adhesive glycoproteins (see Fig. 30-11). Actin filament cross-linking proteins of the plasma membrane skeleton, such as spectrin (see Fig. 7-10) and dystrophin (see Fig. 39-9) are anchored to integral membrane proteins. Relatively little is known about how cells regulate cross-linking proteins, although Ca2+ inhibits binding of some α-actinins to actin.
Adapter Proteins
Eukaryotic cells use multidomain proteins as adapters between signaling pathways and actin assembly (Fig. 33-17). The multidomain protein WASp is defective in the inherited immunodeficiency and bleeding disorder called Wiskott-Aldrich syndrome. The C-terminal domains of WASp (and related proteins N-WASp and Scar/WAVE) activate the Arp2/3 complex to nucleate new actin filaments on the side of existing filaments (Fig. 33-13). Intramolecular interactions autoinhibit WASp. Rho-family GTPases (guanosine triphosphatase), membrane polyphosphoinositides, and polyproline-binding proteins with SH3 domains cooperate to overcome the autoinhibition by binding to parts of WASp involved with the intramolecular interactions. Rac activates Scar by interacting with a complex of regulatoryproteins. An unrelated protein called VASP (vasodilator-stimulated phosphoprotein) also has EVH1 and proline-rich domains as well as an actin-binding domain. These domains allow VASP and related proteins to bind proline-rich ligands in focal adhesions and on the surface of the bacterium Listeria, profilin-actin complexes, and actin filaments. These interactions stimulate actin polymerization in filopodia and “comet tails” assembled by Listeria (see Fig. 37-12).
Functional Redundancy of Actin-Binding Proteins
The diversity and the apparent redundancy of actin-binding proteins are striking. Why should most organisms retain genes for 60 different actin-binding proteins if they have such a limited repertoire of functions: monomer binding, nucleation, capping, severing, cross-linking, stabilizing, and motility? Null mutations show that some proteins are essential for normal physiology. These include myosin-II, Arp2/3 complex, profilin, and cofilin in yeast. On the other hand, organisms can survive genetic deletion of some actin-binding proteins, suggesting that parts of the system are redundant. For example, in a laboratory environment, Dictyostelium tolerates the loss of cross-linking proteins (α-actinin or ABP-120), severin, or one of two profilin genes with only minor defects in behavior and growth. Humans who lack dystrophin develop and grow normally for a few years but later succumb to muscle wasting (see Table 39-1). Mice without their single gelsolin gene reproduce normally, with only mild defects in platelets and other cells.
Actin Dynamics in Live Cells
Cellular actin filaments vary widely in stability. In muscle, the ends of actin filaments exchange subunits over hours. Four proteins stabilize these filaments (see Fig. 39-6). Tropomyosin and nebulin run along the length of the filament, CapZ binds to the barbed end, and tropomodulin binds to the pointed end. These proteins inhibit breakage of the filaments and allow limited exchange of actin subunits at the barbed ends. On the other hand, amoebae and white blood cells protrude and remodel actin-rich pseudopods on a time scale of seconds. Several methods are available to document actin filament dynamics in cells.
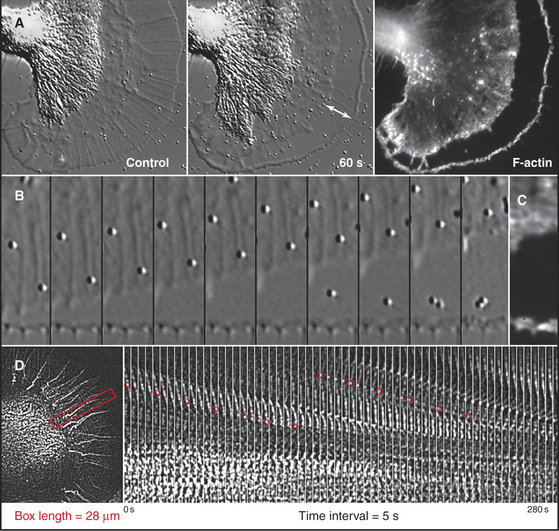
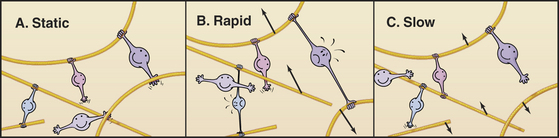
Although light microscopes lack the resolution to observe individual actin filaments crowded together in cytoplasm, assemblies of actin filaments can be imaged directly in favorable cases by DIC or phase contrast microscopy. For example, the parallel arrays of actin filaments in muscle are observed to be very stable, whereas networks of actin filaments in nerve growth cones constantly assemble and move away from the leading edge (Fig. 33-18).
A second approach is to observe the effects of drugs that bind actin such as cytochalasin or latrunculin (Box 33-1). Neither disassembles actin filaments directly, but they both interfere with assembly from monomers, so their effects reveal if filaments are turning over naturally. Muscle actin filaments are relatively resistant to these drugs, but they disrupt the cortical actin filament network in other cells in seconds (Fig. 33-18). When the drug is removed, the cortical actin network reforms rapidly from the leading edge. This response indicates that many cellular filaments turn over rapidly.
BOX 33-1 Tools to Study Actin Filaments: Natural Products Can Stabilize or Destabilize Actin Filaments
Stabilizers Phallotoxins (such as phalloidin) are cyclic peptides that are synthesized by poisonous mushrooms. They bind and stabilize actin filaments by reducing the rate of subunit dissociation to near zero at both ends of the polymer. When introduced into cells by microinjection, phallotoxins inhibit processes that depend on actin filament turnover, including amoeboid movement. They are toxic to humans because they interfere with bile secretion. Fluorescent derivatives of phallotoxins are widely used to localize actin filaments in cells and tissues (Fig. 33-1), as well as to quantify polymerized actin in cells and cell extracts. A sponge toxin, jasplakinolide, has effects similar to phallotoxins.
Actin can also be observed in live cells if labeled with a fluorescent probe. Purified actin can be labeled with a fluorescent dye and microinjected into live cells, where it is incorporated into the actin-containing structures. Expression of actin tagged with green fluorescent protein is even more convenient for following actin in live cells, although caution is required because the bulky green fluorescent protein can interfere with interactions with formins and perhaps other molecules. In muscle, fluorescent actin slowly incorporates, through exchange, at the pointed ends of the filaments. In nonmuscle cells, fluorescent actin is quickly incorporated into all of the filaments. If low levels of fluorescent actin are used, random incorporation into filaments can result in fluorescent “speckles” that can be used to follow the movements and turnover of these subunits (Fig. 33-18D). Figure 38-9 illustrates how bleaching or activating fluorescent actin in a live cell can reveal where filaments assemble and disassemble at the leading edge of motile cells. The associated text explains the biochemical mechanism.
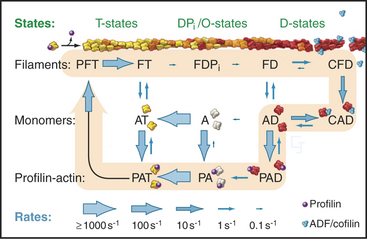
Biochemical and genetic experiments identified the same minimal set of proteins that are essential for maintaining the pool of unpolymerized actin, initiating and terminating new filaments, and controlling disassembly (Fig. 33-19). For example, biochemical reconstitution of actin filament assembly and recycling in the comet tail of the intracellular bacterium Listeria (see Fig. 37-12) requires (in addition to a regulatory protein on the surface of the bacterium) only actin, Arp2/3 complex, ADF/cofilin, profilin, and a capping protein.
The Pool of Unpolymerized Actin
Two complementary mechanisms maintain the pool of unpolymerized actin: monomer binding to profilin and capping of actin filament ends. In vertebrate cells another protein, thymosin-b4, augments the effects of profilin. Thymosin-b4 buffers the free monomer concentration by sequestering actin monomers. Rapid exchange allows monomers to move between profilin and thymosin-b4. The concentrations of profilin and thymosin-b4 exceed the concentration of unpolymerized actin, and these proteins bind tightly enough to reduce the free monomer concentration to the micromolar level. Actin monomers bound to profilin or thymosin-b4 do not nucleate new filaments. However, for profilin to maintain a monomer pool, the barbed ends of most filaments must be capped, since rapid addition of actin-profilin complexes to free barbed ends would quickly deplete the pool of unpolymerized actin. Cells contain enough heterodimeric capping protein, gelsolin, or both to cap the barbed ends of most filaments. Together, monomer binding and capping allow cells to maintain a large pool of actin subunits ready to elongate any barbed ends created by uncapping, severing, or nucleation.
Initiation and Termination of Actin Filaments
A variety of external agonists and internal signals stimulate the conversion of actin from the unpolymerized pool into actin filaments. Examples include the ability of chemoattractants to direct pseudopod formation in slime molds (see Fig. 38-12) and white blood cells (see Fig. 30-13), and the influence of the mitotic spindle on the assembly of the cytokinetic contractile ring (see Fig. 44-22). Polymerization depends on creation of barbed ends, which grow rapidly at rates estimated to be 50 to 500 subunits per second, depending on the concentration of actin-profilin.
Three mechanisms are thought to create free barbed ends: uncapping, severing, and de novo formation of new barbed ends. In many cases, new barbed ends appear to form de novo. At the leading edge of motile cells, small Rho-family GTPases associated with the plasma membrane and polyphosphoinositides activate WASp/Scar proteins, which stimulate Arp2/3 complex to nucleate branches with free barbed ends on the side of older filaments (Figs. 33-2D and 33-13). Formins nucleate filaments for the cleavage furrow and appear to either initiate or sustain the growth of actin filaments in filopodia (see Fig. 38-2A). When thrombin activates platelets (see Fig. 30-14), plasma membrane polyphosphoinositides uncap barbed ends by dissociating gelsolin. Transient increases in the cytoplasmic Ca2+ concentration may also induce gelsolin to sever actin filaments, creating capped barbed ends. Dephosphorylation activates ADF/cofilin proteins, which can sever and nucleate filaments, creating free barbed ends.
Actin Filament Turnover and Subunit Recycling
Actin filaments are long lived if protected by tropomyosin and capping, as in muscle and stress fibers, but many actin filaments, such as those at the leading edge of motile cells, turn over quickly. A possible mechanism involves the hydrolysis of ATP and dissociation of the g-phosphate, reactions that provide a timer to mark older filaments for depolymerization by regulatory proteins (Fig. 33-19). After phosphate dissociates, ADF/cofilin proteins bind ADP-actin subunits in filaments and sever these older filaments. After ADP-actin dissociates from filaments (the details are not yet clear), profilin replaces ADF/cofilin and stimulates the exchange of ADP for ATP. This process recycles actin back to the pool of ATP-actin monomers interacting with profilin. In cells with a high concentration of thymosin-b4, much of the ATP-actin is stored bound to thymosin. Profilin shuttles ATP-actin from this thymosin-b4 buffer to growing filaments. Tropomyosin stabilizes a subset of old filaments by protecting them against ADF/cofilins.
How Do Cells Organize Actin Assemblies?
Cells organize actin filaments in a variety of structures, including cortical networks, microvilli or filopodia, and contractile bundles (Figs. 33-1 to 33-3). Although each cell in a population is unique, all cells of a particular type achieve a similar pattern of organization. How are these patterns specified? Although not yet understood in detail, the mechanisms appear to depend on expression of an appropriate mixture of actin-binding proteins, a prerequisite for self-assembly of particular structures. For example, actin forms bundles similar to microvilli and filopodia when polymerized in the presence of fimbrin and villin, the two major cross-linking proteins found in microvilli. Overexpression of villin in cells induces extension of existing filopodia and formation of new filopodia. Thus, the pool of villin and fimbrin and other components sets the number of microvilli.
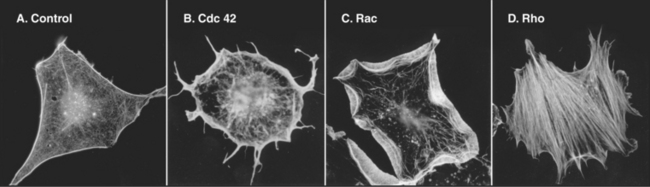
The Rho-family GTPases Cdc42, Rac, and Rho regulate the assembly of many actin filament structures (Fig. 33-20). A complex of proteins including Cdc42 in the bud of yeast cells anchors formins, which mediate the continuous assembly of a cable of actin filaments. The formins anchor the growing barbed ends as the pointed ends extend into the mother cell. These cables are tracks for myosin-V to move cargo into the bud. In motile cells, signals downstream of chemotactic receptors activate Cdc42 and Rac (see Fig. 38-8), which activate WASp/Scar proteins. They in turn stimulate Arp2/3 complex to generate the branched filament network that pushes the membrane forward. Listeria use a surface protein, ActA, to activate Arp2/3 complex, which generates a “comet tail” of actin filaments to push the bacterium through the cytoplasm (see Fig. 37-12). Proteins that mediate endocytosis activate homologs of WASp and other proteins to assemble actin patches in budding yeast (see Fig. 37-11) and fission yeast (Fig. 33-1D).
Physical forces also help to organize actin filaments. Bundles of actin filaments in stress fibers (Fig. 33-1) and the contractile ring during cytokinesis (Fig. 33-3; also see Fig. 44-23) appear to be aligned, at least in part, by tension generated by myosin motors. Activated Rho is required for cytokinesis and also stimulates formation of stress fibers (Fig. 33-20) by activating myosin II. Rho stimulates two kinases that phosphorylate the regulatory light chain and inhibit a phosphatase that reverses the phosphorylation of the light chain (see Fig. 39-21). The success of forces in organizing actin filaments depends on anchoring of the filaments to the plasma membrane—focal contacts in the case of stress fibers (see Fig. 30-11) and the equatorial plasma membrane for the contractile ring (see Fig. 44-23). Cross-linking proteins, such as α-actinin, help to maintain the integrity of these bundles under mechanical stress.
Mechanical Properties of Cytoplasm
Many cross-linking proteins, including α-actinin, have a low affinity for actin filaments with a Kd in the micromolar range. At steady state in vitro, bonds between these cross-linking proteins and actin filaments break and reform on a second or subsecond time scale. Consequently, gels of actin filaments and α-actinin are much more rigid when deformed rapidly than slowly (Fig. 33-21), because cross-links resisting the displacement of the filaments can rearrange if given sufficient time. Dynamic cross-links between filaments allow actin networks to remodel passively as cells move. Cells also remodel the actin cytoskeleton actively by nucleating, severing or depolymerizing filaments.
ACKNOWLEDGMENTS
Thanks go to Charmaine Chan and Aditya Paul for their suggestions on revisions to this chapter.
Amos LA, van den Ent F, Löwe J. Structural/functional homology between the bacterial and eukaryotic cytoskeletons. Curr Opin Cell Biol. 2004;16:24-31.
Bamburg JR, Wiggan OP. ADF/cofilin and actin dynamics in disease. Trends Cell Biol. 2002;12:598-605.
Blessing CA, Ugrinova GT, Goodson HV. Actin and ARPs: Action in the nucleus. Trends Cell Biol. 2004;14:435-442.
Janmey PA, Weitz DA. Dealing with mechanics: Mechanisms of force transduction in cells. Trends Biochem Sci. 2004;29:364-370.
Krause M, Dent EW, Bear JE, et al. Ena/VASP proteins: Regulators of the actin cytoskeleton and cell migration. Annu Rev Cell Dev Biol. 2003;19:541-564.
Kreis T, Vale R, editors. Guidebook to the Cytoskeletal and Motor Proteins, 2nd ed., New York: Oxford University Press, 1999.
Kwiatkowski AV, Gertler FB, Loureiro JJ. Function and regulation of Ena/VASP proteins. Trends Cell Biol. 2003;13:386-392.
Löwe J, van den Ent F, Amos LA. Molecules of the bacterial cytoskeleton. Annu Rev Biophys Biomolec Struct. 2004;33:177-198.
Pollard TD, Blanchoin L, Mullins RD. Biophysics of actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545-576.
Quinlan M, Heuser JE, Kerkhoff E, Mullins RD. Drosophila spire is an actin filament nucleation factor. Nature. 2005;433:382-388.
Stossel TP, Condeelis J, Cooley L, et al. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001;2:138-145.
Wallar BJ, Alberts AS. The formins: Active scaffolds that remodel the cytoskeleton. Trends Cell Biol. 2003;13:435-445.
Wear MA, Cooper JA. Capping protein: New insights into mechanism and regulation. Trends Biochem Sci. 2004;29:418-428.
Weaver AM, Young ME, Lee W-L, Cooper JA. Integration of signals to the Arp2/3 complex. Curr Opin Cell Biol. 2003;15:23-30.
Welch MD, Mullins RD. Cellular control of actin nucleation. Annu Rev Cell Dev Biol. 2002;18:247-288.
Winder SJ. Structural insights into actin-binding, branching and bundling proteins. Curr Opin Cell Biol. 2003;15:14-22.
Zigmond SH. Formin-induced nucleation of actin filaments. Curr Opin Cell Biol. 2004;16:99-105.