Pathology and biology of breast cancer
Introduction
1. to separate patients into groups with significantly differing survival probabilities;
2. to separate patients into groups that include a ‘cured’ group and a group with poor survival;
3. to place a sufficient percentage of cases into each group;
4. to be applicable for all operable breast cancers – small, screen detected as well as symptomatic and young age;
In this chapter common pathological features of breast cancer are reviewed and their role in guiding patients’ management considered.
Traditional factors
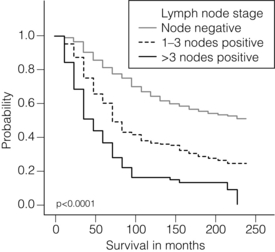
Involvement of local and regional lymph nodes by metastatic carcinoma is one of the most important prognostic factors in breast cancer. The revised TNM staging system for breast cancer has confirmed that the absolute number of axillary lymph nodes involved in metastatic cancer (‘positive lymph nodes’) is one of the most important prognostic factors in breast cancer.1 Lymph node stage has been used consistently as a guide for therapy. However, lymph node stage is considered as a time-dependent factor; the longer the tumour has been growing the more likely it is that lymph nodes are involved by spread. It has also been reported that, when taken alone, lymph node stage is incapable of defining either a cured group or one with close to 100% mortality from breast cancer.
The clinical assessment of nodal status (as in TNM classification) is unreliable.2,3 Palpable nodes may be enlarged because they show benign reactive changes whilst nodes bearing tumour deposits can be impalpable. Although axillary sonography is a moderately sensitive and fairly specific technique in the diagnosis of axillary metastatic involvement,4 histological examination of the lymph nodes from the axilla should be carried out in all patients with primary operable invasive breast cancer. It is well known that patients who have histologically confirmed lymph node involvement have a significantly poorer prognosis than those without nodal metastases. The 10-year survival is reduced from 85% for patients with no nodal involvement to 40% for those with involvement of multiple nodes. Prognosis worsens the greater the number of nodes involved, and the level of nodal involvement can provide useful information; metastasis to the higher level nodes (level II or III) in the axilla and particularly those at the apex (level III) carries a worse prognosis (Fig. 2.1).
Optimal management of the axilla is discussed in Chapter 9.
A refinement of lymph node sampling is provided by the technique of sentinel lymph node (SLN) biopsy. There have been several studies to examine the validity of this technique. The ALMANAC trial provides level I evidence for UK-based practice.3
Intraoperative frozen sections or imprint cytology of axillary lymph nodes have been advocated by some authors. Conventional frozen sections have a high false-negative rate of between 10% and 30%. More intensive intraoperative assessment with serial sections and immunohistochemistry has been described,8 but is time consuming and labour intensive. Frozen section is best applied to selected cases; for example, if the node is macroscopically abnormal and this is confirmed histologically to be metastatic carcinoma, further axillary surgery can be performed immediately. Some studies have found low false-negative rates of 2–3% with intraoperative imprint cytology,9 but not all have been able to achieve this level of accuracy. Lymph node status can be assessed peroperatively using ultrasound combined with FNA or core biopsy, and this reduces the need for preoperative frozen section and imprint cytology to assess axillary nodes intraoperatively.
The detection rate of micrometastases in axillary lymph nodes has been reported to range from 9% to 46%.10 Studies have used serial sectioning with or without immunohistochemical stains to detect micrometastatic foci and these methods have increased detection rates.
The European Working Group for Breast Screening Pathology11 has formulated working guidelines for the assessment and pathological work-up of SLNs in breast cancer. From a literature review available at that time, the committee concluded that it was not possible to determine the significance of micrometastasis or isolated tumour cells. They noted that approximately 18% of cases are associated with other nodal (non-sentinel node) metastases. False-negative rates are most often determined via immunohistochemistry (IHC). However, at present it is recommended that an intensive work-up is not justified on a population level. The committee did suggest multilevel assessment and, where resources permit, intraoperative assessment, although the results of Z0011 (see Chapter 7) suggest that some node-positive patients may not require axillary dissection and this brings the whole issue of intraoperative assessment into question. The current view is that IHC should not be used routinely to assess axillary lymph nodes.
Tumour size
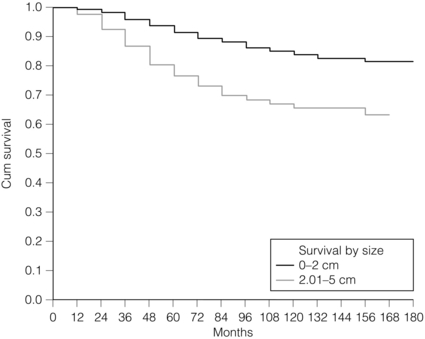
Tumour size is one of the most powerful predictors of prognostic factor in breast cancer.12,13 The frequency of nodal metastases in patients with tumours < 10 mm is 10–20%,13 and node-negative patients with tumours < 10 mm have a 10-year disease-free survival rate of greater than 90%.14 Tumour size is a time-dependent prognostic factor that depends on the period between tumour development and detection, and on the balance between tumour cell proliferation and death (tumour growth rate). It is well known that the association between increasing tumour size and increasing number of positive lymph nodes and worse outcome is highly statistically significant.13 Therefore, the main aim of population screening with mammography is to detect smaller tumours that are likely to have a better outcome than those that present symptomatically (of a larger size). Patients with smaller tumours have a better long-term survival than those with larger tumours (Fig. 2.2). Estimation of tumour size has assumed particular importance since the introduction of population screening. In most studies the frequency of axillary lymph node metastasis in small < 15 mm (so-called minimally invasive carcinoma (MIC)) invasive carcinomas is 15–20%,15–17 compared with over 40% in tumours measuring 15 mm or more.18 During the prevalent round of breast screening even more favourable results are obtained, with the frequency of axillary lymph node metastasis ranging from 0% to 15%.19–22 The Nottingham Tenovus Primary Breast Cancer Study (NTPBCS) has generated data that suggest the cut-off point of 10 mm is not the best discriminator for MIC. Life-table analysis of survival curves found no difference between tumours measuring up to 9 mm and those measuring 10–14 mm. This indicates that < 15 mm may be a more realistic watershed in defining small, invasive, carcinomas of good prognosis. It is clear that pathological tumour size is a valuable prognostic factor and it has become an important quality assurance measure for breast screening.21,23–26 It is also used in part to judge the ability of radiologists to detect small impalpable invasive carcinomas.
Differentiation
Certain histological types of invasive carcinoma carry a favourable prognosis. Tubular, mucinous, invasive cribriform, medullary and tubulolobular types, together with rare tumour types such as adenoid cystic carcinoma, adenomyoepithelioma and low-grade and squamous carcinoma, have all been reported to have a more favourable outcome than invasive carcinoma of no special type (ductal NST). Undoubtedly, assessment of histological type provides prognostic information in breast cancer. However, this effect is relatively small in multivariate analysis27 when compared with the prognostic value of histological grade; histological type may prove to be more useful in increasing our understanding of the biology of breast cancers.28
Tumour grade
The Nottingham method (Elston and Ellis) is a modification and improvement of previously existing morphological grading systems and is able to provide greater objectivity of grade.30
• Grade 1 – well differentiated = score 3–5 points.
It should be noted that grade is valuable irrespective of histological type.
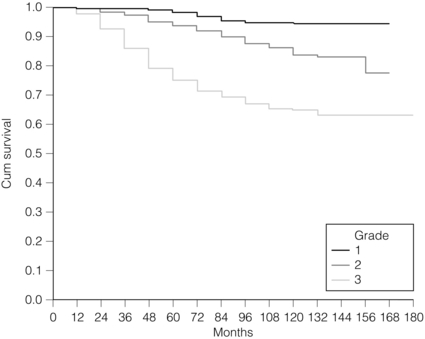
There is a highly significant correlation between tumour grade with long-term prognosis (Fig. 2.3); patients with grade 1 tumours have a 93% chance of surviving 10 years after diagnosis, whereas in patients with grade 3 tumours this is reduced to 70%. It has now been shown conclusively that the Nottingham method, with its more objective criteria, has excellent reproducibility when used by experienced pathologists.
Vascular invasion
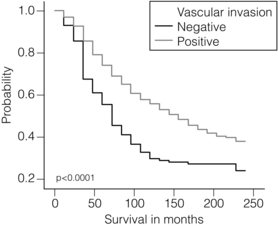
The presence of tumour emboli in vascular and lymphatic spaces has emerged as an important prognostic factor. Several studies have now shown that the presence of vascular invasion correlates closely with local and regional lymph node involvement.33,34 It has been suggested that it can provide prognostic information as powerful as lymph node stage. Reproducibility is the limiting factor in its widespread adoption and routine clinical assessment. In a Nottingham study of assessing vascular invasion, the issue of reproducibility was addressed specifically. Of 1704 cases, a subset of 400 cases was examined by two or more pathologists. There was a 77% overall inter-observer agreement on histological features and an 85.8% overall agreement in the classification of vascular invasion. Other studies have reported a similarly high concurrence between pathologists.35,36 The assessment of vascular invasion is subjective but there is good evidence that a high rate of concurrence can be obtained as long as strict criteria are used. Lymphatic and vascular invasion is considered to be a valuable surrogate for lymph node stage in cases where nodes have not been removed for examination. In patients whose axillary nodes are tumour free on histological examination there is a correlation between the presence of lymphovascular invasion (LVI) and early recurrence.
Molecular/predictive factors
Oestrogen receptors (ERs)/progesterone receptors (PRs)
Knowledge of the expression of hormone receptors and their subcellular regulatory pathways is one of the classic examples of the use of translational medicine in breast cancer to further diagnosis and treatment of this disease. The degree of ER expression is used to predict an individual’s response to hormone therapy. Both ERs and PRs are steroid receptors that are located in the cell nucleus. Oestrogen and progesterone are considered to diffuse into cells, or be transported to the nucleus. Genes, regulated by steroid receptors, are involved in controlling cell growth, and it is currently believed that these effects are the most relevant to ERs and that ER expression influences the behaviour and treatment of breast cancer.38 Elucidation of downstream effect on genes that are known to be influenced by hormones has led to the inclusion of these ER-regulated genes in high-density oligonucleotide array panels.39 This may better define those pathways that are endocrine responsive and may lead to improved therapies with reduced side-effects and a greater potential for cure.40
In unselected patients, approximately 30% will respond to endocrine therapy. A tumour with both ER and PR expression has a 78% chance of responding to hormone therapy while those cancers that are ER/PR negative respond rarely, if ever.41–43 Due to their close relationship with histological grade, ERs are not of independent prognostic significance.43
Methods of measuring ERs and PRs
There are a multitude of methods available; however, in clinical practice most are based on two distinct strategies. The first is an older method based on ligand-binding methods, i.e. radiolabelled steroid ligand is used to detect the receptor. The second relies on the recognition of the receptor protein by specific antibodies. Several studies examined the correlation between assay and outcome/response and studies have suggested that IHC analyses have a number of advantages.42 IHC is currently the most commonly used method because it can be performed on paraffin-embedded material and allows assessment of the expression specifically in either invasive or in situ cancer.
Type 1 growth factor receptors
EGFR
Methods of testing: Several techniques directed at DNA, RNA, protein (and functionally active (activated) as well) or serum can be employed to identify EGFR expression in breast cancer.
Of all such methods, IHC is the most practical. The advantages of IHC in EGFR as in HER-2 testing include reproducibility, ease of interpretation and the low cost compared to other techniques. It can also be employed on archival tissue. Protein levels can be quantified by Western analysis or enzyme immunoassay; however, architecture of tissue is lost in these procedures and there may be contamination by normal tissue cells or ductal carcinoma in situ. Serum EGFR levels have been examined in breast carcinoma; however, few data are available.44,45 Due to the paucity of relevant data, it is difficult to draw conclusions on the current value of measuring EGFR in the serum of patients.
Prognostic and predictive value: The prognostic value of EGFR in breast cancer has been examined in several large studies. There remains no consensus on its prognostic value. The lack of a clear picture has been attributed to several factors: lack of a standard assay (monoclonal vs. polyclonal), lack of a cut-off for positivity and great variation in study designs (size, follow-up and influence of adjuvant therapy). Klijn et al.46 reviewed data from over 5000 patients where EGFR had been assessed. Findings from this review demonstrated a great heterogeneity of study design, levels of cut-offs for positive and negative, and, not surprisingly, differences in its prognostic value in these various studies.
The predictive value of this marker for hormone resistance or responsiveness is better defined. EGFR tumours are considered more resistant to endocrine therapy, and there exists an inverse relationship with EGFR-negative cancers (they are more likely ER positive), being more often sensitive to endocrine manipulation. Though some studies have been able to demonstrate that EGFR status is a predictor of tamoxifen failure and even response rates,50 there are conflicting data from well-designed level II studies51 that have shown no value of EGFR in predicting the efficacy of tamoxifen in high-risk postmenopausal women.
HER-2
Methods of HER-2 testing: Current guidelines for HER-2 testing52 specify the methods that are suitable to detect either the HER-2 protein (by IHC) or gene amplification (using FISH or other in situ methods). Guidelines stress the need for stringent, reproducible and consistent criteria for testing.
Immunohistochemistry for HER-2 testing: Among the methods in use for determining HER-2 status, IHC is the most widely used. In studies employing various commercially available antibodies, a wide variety of sensitivity and specificity in fixed paraffin-embedded tissues is seen.53,54 Antigen retrieval techniques are currently not standardised and they introduce the potential for false-positive staining. Nonetheless, IHC possesses many advantages to support its widespread adoption: (i) it allows for the preservation of tissue architecture and so can be used to identify local areas of overexpression within a heterogeneous sample, and can distinguish between HER-2 positivity in in-situ and invasive cancer; (ii) it is applicable to routine patient samples, facilitating use as a diagnostic test, and this allows prospective and retrospective research studies of HER-2 status to be undertaken.
Two Food and Drug Administration (FDA)-approved IHC tests for determining HER-2 status are available: HercepTest (DAKO, Carpeteria, CA, USA), based upon a polyclonal antibody; and CB11 (Pathway, Ventana Medical Systems, Tucson, AZ, USA), based upon a monoclonal antibody. The National Comprehensive Cancer Network guidelines55 classify an IHC score of 0 or 1 + as representing HER-2-negative status, 3 + as positive, while 2 + is equivocal. Positive staining is defined as strong, continuous membranous expression of HER-2 in at least 10% of tumour cells. However, a joint report from the American Society of Clinical Oncology (ASCO) and the College of American Pathologists (CAP)52 specified a threshold of > 30% strong circumferential membrane staining for a positive result. If both uniformity and a homogeneous, dark circumferential pattern are seen, resultant cases are likely to be amplified by FISH as well as positive for HER-2 protein expression. The equivocal range for IHC (score 2 +), which may include up to 23% of samples, is defined as complete membrane staining that is either non-uniform or weak in intensity but with obvious circumferential distribution in at least 10% of cells. Equivocal or inconclusive results should be tested by FISH. Consistent with previous guidelines, a negative HER-2 test is defined as either an IHC result of 0 or 1 + for cellular membrane protein expression (no staining or weak, incomplete membrane staining in any proportion of tumour cells), though up to 5% of 1 + on IHC may be FISH positive.
In situ hybridisation for HER-2 testing: FISH and CISH measure directly the number of HER2 genes per chromosome 17, and when there is a chromosome centromeric enumeration probe (CEP) included, the copy number of chromosome 17 gene amplification is defined as an increase in HER-2/CEP17 ratio above 2.0. ISH results are semiquantitative, counting the number of signals in non-overlapping interphase nuclei of the lesion using either single-colour (HER-2 probe only, e.g. Ventana Inform) or dual-colour hybridisation (using HER-2 and chromosome 17 centromere probes simultaneously (e.g. Abbott, Chicago, USA, DAKO, Copenhagen, Denmark, etc.), the latter making it easier to distinguish true HER-2 amplification from chromosomal aneuploidy. ISH allows simultaneous morphological assessment, where evaluation of gene amplification can be restricted to invasive carcinoma cells. Many studies have compared FISH and IHC in the evaluation of HER-2 and have demonstrated concordance between the two techniques of up to 91%. Two studies have shown that FISH predicts more accurately HER-2 positivity than IHC when applied to molecularly characterised breast cancers.56,57
Recommended guidelines for HER-2 assessment in the UK have recently been updated and the reader is referred to these guidelines for further details.58
In brief, a two-phase testing algorithm based on IHC assay as the primary screen with FISH being reserved for equivocal cases is currently recommended. This is based on evidence showing very good concordance between IHC and FISH results on breast carcinomas from 37 laboratories when tested in experienced reference centres.59
Prognostic significance and association with other prognostic factors: The seminal work by Slamon et al.60 in 1987 showed that HER2 gene amplification independently predicted overall survival (OS) and disease-free survival (DFS) in a multivariate analysis in node-positive patients. Since then most large studies have confirmed this relationship in multivariate analysis. It is now well established that there is a significant correlation between HER-2 overexpression/amplification and poor prognosis for patients with nodal metastasis. There was until recently no consensus on the prognostic value of HER-2 in node-negative breast cancer patients, a group most often diagnosed through screening and representing a subgroup that could potentially benefit from appropriate adjuvant therapy. More recent studies have, however, shown a consistently poorer prognosis when comparing HER-2-positive versus -negative small node-negative breast cancers.
Rilke et al.61 reported on the prognostic significance of HER-2 expression and its relationship with other prognostic factors. Using specimens from 1210 consecutive patients treated between 1968 and 1971 at a single institution (National Cancer Institute of Milan), with no systemic adjuvant therapy and 20-year follow-up, they found overexpression of HER-2 in 23% and showed a negative impact on survival of node-positive but not node-negative patients. Analysis of HER-2 in relation to the presence of lymphoplasmacytic infiltrate (LPI; favourable prognosis) and nodal status demonstrated that in node-negative, LPI-negative patients, HER-2 overexpression showed the same level of correlation with poor prognosis as with those patients with nodal metastasis. However, in the patients with node-negative disease and LPI positivity, HER-2 overexpression correlated with good prognosis. Some studies have reported a prognostic value for HER-2 in node-negative patients in selected subgroups,62–64 whereas others have shown no correlation.65,66
Mirza et al.66 have published a systematic review of prognostic factors in node-negative disease. Data for HER-2 showed a lack of standardisation of assays and no association with survival. However, HER-2 status was shown to be of independent prognostic significance in several large studies with long-term follow-up, with worstened prognosis for those node-negative patients with HER-2-positive signalling.67,68,69
There is at present no agreement on the association between HER-2 and other prognostic factors. Several studies have shown a lack of association between HER-2 status and tumour size,60,70,71 although some do report a correlation.61,66,72–76
Prediction of response to therapy: hormonal therapy: Transfection of normal breast cancer cells with the HER2 gene has been shown to result in acquisition of oestrogen-independent growth that is insensitive to tamoxifen.77,78
A number of clinical studies, using various end-points such as time to relapse, more rapid spread to other sites, and DFS or OS, have reported an association between HER-2 positivity and resistance to hormonal therapy.79–83 Some reports have described specific resistance to tamoxifen in HER-2-overexpressing tumours.81,82 The 20-year update of the Naples GUN Trial81 found that HER-2 overexpression not only predicted resistance to tamoxifen, but that HER-2-positive patients had a worse outcome on tamoxifen therapy compared with those who were untreated.
Several studies have also shown a reduction in response rates to hormonal therapy. Metastatic breast cancer that overexpressed HER-2, measured by high plasma levels of extracellular domain, demonstrated a substantial reduction in response rate to hormonal therapy.84 Other studies have failed to find an association or even a trend between HER-2 status and response to hormonal therapy.85–87 Elledge et al.87 examined the response to tamoxifen in 205 tumours with ER-positive disease. In HER-2-positive compared to HER-2-negative patients, they found no significant evidence for a poorer response, time to treatment failure or survival. In a more recent study, the relationship between HER-2 overexpression and response to tamoxifen was examined in the adjuvant setting in 741 (650 ER positive, 91 ER negative/PR positive) of the total of 1572 patients in the CALGB 8541 Trial who had HER-2 measured.88 Tamoxifen significantly improved response, DFS and OS, irrespective of HER-2 status. However, tamoxifen was not randomised within this trial and all patients received one of three regimens of doxorubicin. Thus these data on tamoxifen resistance have limitations to their interpretation.
With regards to HER-2 and prognosis, not only is there clinical value in its positive expression but also in its absence – in so-called ‘triple-negative phenotype’ cancers (TNP), i.e. cancers that are HER-2, ER and PR negative. It also denotes a biologically different subgroup. The recognition of basal phenotype in these triple-negative cancers is of growing importance, with several lines of evidence supporting the view that triple-negative tumours and basal phenotype are not interchangeable but rather distinct entities. Ninety-one per cent of TNP tumours display a significant association with the basal-like centroid and about 20% of non-TNP tumours cluster together with TNP tumours in the ‘basal-like’ cluster; these data support the view that the majority of, but not all, TNP tumours have a basal-like phenotype and that the majority of, but not all, basal-like tumours have a TNP phenotype.89 Triple-negative breast cancers have a relapse pattern that is very different from hormone-positive breast cancers: the risk of relapse is much higher for the first 3–5 years but drops sharply and substantially below that of hormone-positive breast cancers after that. This relapse pattern has been recognised for all types of triple-negative cancers for which sufficient data exist, although the absolute relapse and survival rates differ across subtypes.
Previous studies have shown that the expression of ‘basal markers’ (i.e. CK5/6, CK14, CK17 and/or EGFR) is associated with a poor prognosis, regardless of hormone receptor expression. The expression of basal markers (basal cytokeratins and EGFR) in TNP tumours (core basal phenotype) also correlates with a worse prognosis and identifies a clinically distinct subgroup within the TNP group. Moreover, it should be noted that identification of a subgroup of tumours solely based on the lack of expression of immunohistochemistry (for example, TNP) risks misassignment based on technical artefacts.89
An in-depth discussion of these issues is beyond the scope of this work and the reader is directed to detailed reviews by Rakha et al.89
Histopathology of patients with BRCA1 and BRCA2 mutations
The cancers that develop in patients with a genetic predisposition to breast cancer as a result of mutations in the breast cancer susceptibility genes BRCA1 and BRCA2 are of great clinical interest. Identification of histological features that could indicate a genetic predisposition would be useful in providing an insight into the function of these genes and may aid in identifying those in whom screening for genetic mutations would be useful. There is a general agreement that BRCA1-related cancers are more frequently ‘medullary-like’ carcinomas and high grade compared to those in patients without this genetic alteration. Cancers associated with BRCA1 mutations have a significantly higher mitotic rate, a larger proportion of tumour with a continuous pushing margin and more lymphocytic infiltration than sporadic breast cancers.90,91 These are also likely to be less positive for ERs and PRs and are more often aneuploid, have a high S-phase fraction, show greater accumulation of p52 protein and expression of basal cytokeratins (CK5/6, CK14 and CK17) than do sporadic breast cancers.92–97 However, none of these features alone or in combination can be used to identify a cancer as being from a BRCA1 gene mutation carrier. Pathological features reported in BRCA2-related breast cancer have been less consistent. Marcus et al.92 noted a significantly higher proportion of tubulolobular cancers in BRCA2 mutation carriers than in other patients. However, it has been reported in another study that tubular carcinomas are less common in BRCA2 mutation carriers.94 Armes et al.97 investigated the histological phenotypes of breast carcinoma in women under 40 years of age with and without BRCA1 and BRCA2 germ-line mutations. It was found that the pleomorphic variant of invasive lobular carcinoma was more common in patients with BRCA2 mutations. Others have reported that BRCA2-related cancers tend to be of high histological grade,94,98 whereas others have not noted any significant difference in grade between BRCA2-related cancers and controls.99
High-throughput molecular techniques as prognostic and predictive tools in breast cancer
Mechanisms of genetic aberrations
The genetic fingerprint provides information on normal cellular processes and morphological/phenotypic expression. When this message is altered, it forms the nidus for the development and progression of cancer. These alterations can be caused by DNA mutations, chromosomal aberrations, epigenetic modification and protein interactions. DNA mutations can lead to a change in gene function. These mutations are often due to base substitutions that may directly cause a stop codon. As a result the gene is only partly transcribed and any functional protein production terminated. Chromosomal instability can be due to DNA modification, mutation or viral genome integration. Changes in chromosome numbers are also seen and cancers can be aneuploid (60–90 chromosomes) or near diploid (46 chromosomes). In solid tumours virtually all chromosomal rearrangements are unbalanced, the net result being loss or gain in certain parts of the chromosome. By screening the genome, current losses and gains can be identified. This information can be used to investigate regions of overlap to reveal the genes that are involved in malignant transformation and cancer progression. In some situations, the DNA copy number and code remain intact but the accessibility of this DNA for transcription is affected by epigenetic modifications. This process can occur through DNA methylation. It directly silences genes and interferes with the binding of transcription factors by changing the chromatin structure around the gene. Global hypomethylation is a characteristic feature of the genome of a cancer cell. However, some sequences can be hypermethylated, such as CpG islands, which tend to lie around the transcription start sites of approximately half of all human genes. DNA hypomethylation has been shown to correlate with chromosome instability. Chromatin architecture remodelling is essential for gene transcription; thus the prognostic significance of quantitative chromatin changes.100,101
Assessment of genomic status in breast cancer
Several high-throughput molecular techniques have been developed to assess the status of the genome in a given cell population, in terms of copy number, DNA sequencing and structure. Large-scale DNA or gene copy number alterations can be assessed using loss of heterozygosity (LOH) and comparative genomic hybridisation (CGH) studies as well as other techniques. LOH can be used to identify chromosomal regions where allelic losses are more frequent. In one study,102 a panel of 150 polymorphic microsatellite markers from throughout the whole genome was used to identify such regions. These data were correlated with clinicopathological features and showed four specific loci that correlated with lymph node metastasis (11q23–24, 13q12, 17p13.3 and 22q13). Microsatellites can also be used to check the ability of cells to repair DNA replication errors. Microsatellite instability (MSI) can be employed to identify both genetic and epigenetic modification. It has been shown that both genetic and epigenetic alterations of hMSH2 and hMLH1 contribute to genomic instability and tumour progression in sporadic breast cancer.
Chromosome-based CGH can be used to identify the loss of one or both copies of a given gene, as well as regions of amplification. In addition, the technique provides information on the number of copies of any part of each chromosome throughout the whole genome. There have been several studies using CGH in breast carcinoma,103,104 and those with clinical follow-up have led to the identification of genetic changes related to prognosis. However, the resolution of every chromosome-based CGH technique was limited to approximately 10 million base pairs (Mbp). This limitation makes it difficult to link copy number changes to the genes involved. Another limitation to this technique is the need to perform karyotyping for target identification in every experiment.
Therefore, array CGH has been developed to overcome the problems of chromosome-based CGH. Array CGH does not require a normal metaphase spread but rather an array of DNA fragments (100 bp to 100 kb) and their precise chromosomal locus. This approach can provide resolutions up to 1 Mb. Albertson et al. utilised this technique to map the recurrent breast cancer amplicon at chromosome 20q12.3. They were able to demonstrate that what was previously described as a single amplicon is in fact two distinct amplicons (ZNF217 and CYP24). A novel amplicon at 17q21.3 that is implicated in the amplification and overexpression of the HOXB7 gene in breast cancer has also been recently characterised using array CGH.105
Assessment of gene expression in breast cancer
In breast cancer, the class discovery studies were pioneered by the Stanford group,106 which proved the principle that breast cancer could be classified into molecularly distinct groups based upon gene expression profiles and their similarity to normal cell counterparts. Multiple independent studies have confirmed and expanded the original results. According to these studies, breast cancer was classified into two main classes (ER-positive and ER-negative tumours) and each one can be classified into multiple molecularly distinct subclasses. For example, the ER-negative tumours encompass three subgroups, one overexpressing HER-2, one with tumours expressing genes characteristic of basal/myoepithelial cells (basal-like cancer), and one with a gene expression profile similar to normal breast tissue. cDNAs have also been used to distinguish cancers with BRCA1/BRCA2 mutations107,108 and to determine ER status,109 lymph nodes status110,111 and prognostic subgroups in node-negative breast cancer.112 These recent studies demonstrate the ability of cDNA to have a direct translational use in clinical practice.
Proteomics
These global expression profiling approaches yield candidate proteins and related genes that require verification through application of other techniques. Tissue microarrays (TMAs) provide a method for high-throughput protein expression analysis of large cohorts of archival samples that can be readily linked to clinicopathological and long-term follow-up databases. TMAs have expedited the validation of the prognostic and predictive significance of several candidate biomarkers. The technique allows for a composite slide of up to 1000 cores of tissue to be constructed into one paraffin block. The advantages are obvious in screening for novel protein expression. There are concerns on the validity of the cores as being representative samples and indeed this aspect has not been well studied, with only one study examining a large sample size with correlation to corresponding larger histology sections.103 This technology and its application have been extensively reviewed.113
Clinical use of the high-throughput molecular techniques
It is envisaged that gene expression profiles may be able to guide decisions on the choice of hormonal, chemotherapeutic or targeted agents for each individual in the future. Presently, one of the best examples of the use of genomics and proteomics in clinical practice is the use of HER-2 expression/ amplification to select patients for trastuzumab. Such profiles are useful to identify prognostically significant genes, which studies have demonstrated can perform better than traditional markers.104,114 In metastatic disease, expression patterns can be used to establish the origin of the metastatic lesion in patients with more than one primary. Transcription profiling has also been shown to differentiate accurately breast cancers with germ-line mutations in BRCA1/BRCA2 from those without such mutations. Such findings, if validated in other studies, open the way for molecular phenotyping in high-risk families. In addition, gene expression profiling has been used to develop expression predictors (class prediction studies) that can be used for many types of clinical management decisions, including risk assessment, diagnostic testing, prognostic stratification and treatment selection. There are two types of class prediction studies in breast cancer: prognostic and predictive class prediction. Prognostic class prediction includes (i) poor-prognosis gene signatures that can discriminate between a good and a poor outcome by comparison between highly aggressive and less aggressive primary tumours, and (ii) recurrence score gene signatures that define tumours based on the risk of disease relapse. The predictive class prediction includes predictors of response to therapy.115,116
An example of a prognostic class prediction study is that reported by van’t Veer et al. Expression profiles of 117 primary breast cancers were compared with known prognostic factors and matched with 5 years of follow-up data. Twenty-five thousand genes were used to generate expression profiles, which separated the tumours into two groups. Group 1 developed distant metastasis in 34% and in group 2 70% developed distant metastases. From the 25 000-gene set, 70 genes had great accuracy in predicting recurrent disease. Multivariate analysis with grade, size, LVI, age and ER showed the poor-prognosis microarray profile to be an independent predictor of recurrent disease. This approach was further tested in 295 patients and again the use of gene profiling was able to accurately identify a poor-prognosis group.101,109 MammaPrint® assay, which is based on the Amsterdam 70-gene gene signature, is currently used as a molecular diagnostic test for breast cancer prognostication and prediction. Another predictor calculates a recurrence score on the basis of the expression of 21 known genes, with the use of RT–PCR in formalin-fixed, paraffin-embedded tissue. The ‘Oncotype DX assay’ has been developed by researchers at Genomic Health. Oncotype DX is currently used as a diagnostic test to quantify the likelihood of disease recurrence in women with early-stage breast cancer and assesses the likely benefit from tamoxifen and certain types of chemotherapy. Oncotype DX employs a mathematical algorithm called the recurrence score (range 1–100) to calculate continuous risk for relapse and death for patients receiving adjuvant tamoxifen, and has recently been shown in a large population-based case–control study to be an effective predictive test for ER-positive, node-negative breast cancer patients treated or untreated with tamoxifen and no chemotherapy.117 In addition, a number of microarray studies have been published which identified other prognostic signatures with clinical significance. For example, Change et al.118 have used a ‘wound-response gene expression signature’ to stratify breast cancer patients based on the hypothesis that features of the molecular programme of normal wound healing might play an important role in cancer metastasis. They found that this gene signature can improve the risk stratification of early breast cancer over that provided by standard clinicopathological features. A gene expression signature of hypoxia response, derived from studies of cultured mammary epithelial cells’ ‘hypoxia gene signature’, showed a strong predictor of clinical outcomes in breast cancer.119
Although proteomics has a long history that predates profiling at the RNA level, difficulties in protein identification and the lack of reproducibility of several assay platforms have limited the utility of such profiling systems. In breast cancer research, however, proteomics has begun to take on a role in the monitoring of response, prediction of resistance and relapse in patients treated with novel bio-directed therapies.120
Clinical use of prognostic factors in patient management
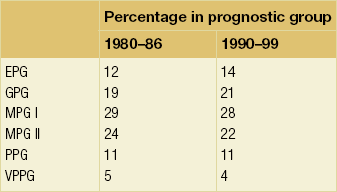
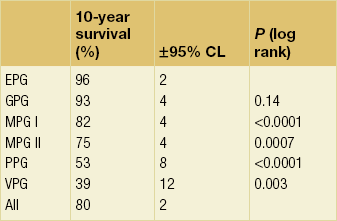
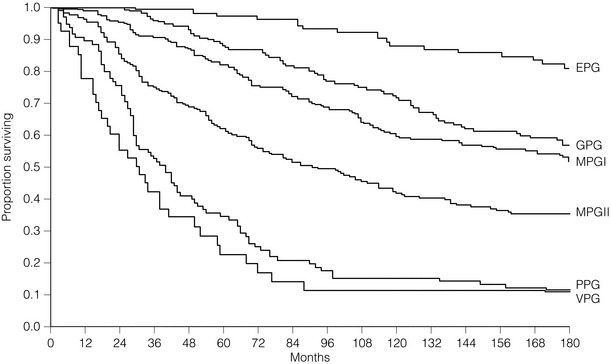
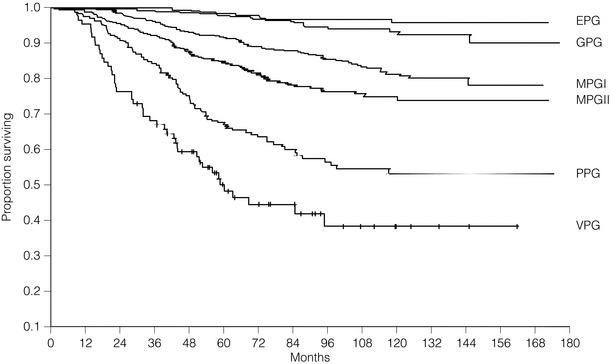
Arbitrary cut-off points of 3.4 and 5.4 are used to divide patients into six prognostic groups: excellent (EPG), good (GPG), moderate (MPG) I and II, poor (PPG) and very poor (VPG) (Tables 2.1–2.3 , Figs 2.5 and 2.6).
Table 2.3
Relative risk reduction by NPI
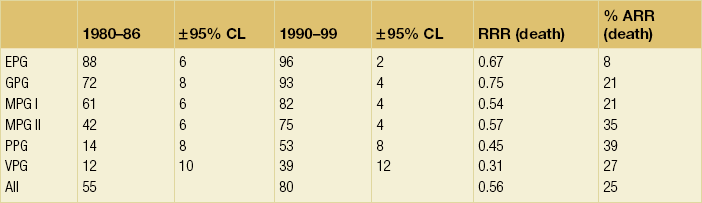
ARR, absolute risk reduction; CL, confidence limit; RRR, relative risk reduction.
The NPI provides extremely powerful prognostic information within the NTPBCS and was able to demonstrate its utility and reproducibility in studies from other centres. In this respect Henson et al.,18 in a retrospective analysis of prognostic data in over 22 000 women from the SEER Programme of the National Cancer Institute in the USA, confirmed that a combination of lymph node stage and histological grade improved prediction of prognosis. In a similar way to the NTPBCS, Chevallier et al.121 have identified ‘young’ age, tumour size and histological grade as factors to be added to lymph node stage in the prediction of recurrence; these factors were used to divide lymph node-negative patients into three prognostic groups.
One of the strengths of the NPI is the fact that it has been verified prospectively in the NTPBCS. Further confirmation of its value has been provided by its validation in two large multicentre studies involving nearly 11 000 patients in total.122,123 Such studies demonstrate the inherent power of the pathological factors used in the NPI, which has become the most widely used index for the management of patients with breast cancer, certainly in the UK.
Other pathology-based prognostic indices used in breast cancer include Adjuvant!Online (AO)124 and the St Gallen criteria,125 and more recently Predict (a new UK prognostic model that predicts survival following surgery for invasive breast cancer).126 This tool is a population-based validation of the prognostic model PREDICT for early breast cancer.
Adjuvant!Online (http://www.adjuvantonline.com) was developed in the USA and published in 2001; users can input information on a patient’s age, ER status, tumour grade, tumour size and number of positive nodes, and obtain predictions of 10-year OS (the likelihood of being alive 10 years after the diagnosis of breast cancer was first carried out), BCSS (the likelihood of not dying of breast cancer within 10 years of diagnosis) and event-free survival (EFS; the likelihood of surviving 10 years without recurrence (local, regional or distant), a second primary breast cancer, or death from breast cancer), both with and without any proposed adjuvant therapy. The performance of AO has been evaluated in cohorts of patients in Germany, Canada and the UK. It has an advantage over the NPI in that it integrates treatment, and for this reason AO is the programme most commonly used by oncologists.
All of the above discussion applies to primary operable breast cancer. Prognostic factors have also been explored in locally advanced and metastatic breast cancer, albeit not to the same extent. In a small study of locally advanced pancreatic cancer patients (n = 60) treated with either tamoxifen or radiation therapy and with crossover upon failure, response to therapy correlated significantly with histological grade (P = 0.02), ER (P = 0.02), PR status (P = 0.02), mitotic index (P = 0.01) and tumour ploidy (P = 0.04). Survival from initial therapy correlated significantly with ER (P = 0.01) and PR status (P = 0.04).127 In these patients, histological grade, mitotic index, tumour ploidy, and ER and PR status of the primary tumour may predict response and prognosis.
These authors also explored the prognostic value of certain factors as a composite index to guide decision-making in metastatic disease.128 The advanced breast cancer (ABC) index comprises the grade, ER status, site of initial metastasis and disease-free interval. This index was tested both retrospectively and prospectively and shown to separate groups where the survival between each was highly significant (P < 0.001).
Conclusion
Modern management of breast cancer requires a multifaceted understanding of both the clinicopathological features and the molecular portraits of the cancer. Emerging evidence is suggesting that genomic approaches to prognosticating patients can be complementary to standard prognostic tools. Existing combinations of such factors are either largely clinical (e.g. NPI) or purely molecular (e.g. Oncotype DX), but a hybridised index is likely to yield more information. Updating the traditional NPI and incorporating well-established prognostic factors such as grade, stage and tumour size with biological factors have produced NPI+. This new tool, which combines updated methods for assessment of histological grade and tumour stage with biological class determination, has recently been shown to provide significant prediction of outcome in all biological classes.129
Contents of the final surgical pathology report: the minimum dataset
The Royal College of Pathologists minimum dataset has been approved by the NHS Breast Screening Programme and the European Commission Working Group, the British Association of Surgical Oncologists, the British Breast Group and the United Kingdom Association of Cancer Registries (Fig. 2.7).
References
1. Singletary, S.E., Allred, C., Ashley, P., et al, Revision of the American Joint Committee on Cancer staging system for breast cancer. J Clin Oncol 2002; 20:3628–3636. 12202663
2. Rampaul, R.S., Mullinger, K., Macmillan, R.D., et al, Incidence of clinically significant lymphoedema as a complication following surgery for primary operable breast cancer. Eur J Cancer. 2003;39(15):2165–2167. 14522373
3. Mansel, R.E., Fallowfield, L., Kissin, M., et al, Randomized multicenter trial of sentinel node biopsy versus standard axillary treatment in operable breast cancer: the ALMANAC Trial. J Natl Cancer Inst. 2006;98(9):599–609. 16670385
4. Alvarez, S., Anorbe, E., Alcorta, P., et al, Role of sonography in the diagnosis of axillary lymph node metastases in breast cancer: a systematic review. Am J Roentgenol 2006; 186:1342–1348. 16632729
5. Rampaul, R.S., Evans, A.J., Ellis, I.O., et al. Long term regional recurrence and survival after axillary nodal sampling for breast cancer. Eur J Cancer. 2003; 1(4):23. [(abstr.)].
6. Shukla, H.S., Melhuish, J., Mansel, R.E., et al, Does local therapy affect survival rats in breast cancer? Ann Surg Oncol. 1999;6(5):455–460. 10458683
7. Macmillan, R.D., Rampaul, R.S., Lewis, S., et al, Preoperative ultrasound-guided node biopsy and sentinel node augmented node sample is best practice. Eur J Cancer. 2004;40(2):176–178. 14728929
8. Veronesi, U., Galimberti, V., Zurrida, S., et al, Sentinel lymph node biopsy as an indicator for axillary dissection in early breast cancer. Eur J Cancer. 2001;37(4):454–458. 11267853
9. Veronesi, U., Paganelli, G., Viale, G., et al, Sentinel lymph node biopsy and axillary dissection in breast cancer: results in a large series. J Natl Cancer Inst. 1999;91(4):368–373. 10050871
10. Schwartz, G.F., Guiliano, A.E., Veronesi, U., Consensus Conference Committee, Proceeding of the consensus conference of the role of sentinel lymph node biopsy in carcinoma or the breast April 19–22, 2001, Philadelphia, PA, USA. Breast J. 2002;8(3):124–138. 12078657
11. Cserni, G., Amendoeira, I., Apostolikas, N., et al, Discrepancies in current practice of pathological evaluation of sentinel lymph nodes in breast cancer. Results of a questionnaire based survey by the European Working Group for Breast Screening Pathology. J Clin Pathol. 2004;57(7):695–701. 15220360
12. Fisher, E.R., Fisher, B., Sass, R., et al, Pathologic findings from the National Surgical Adjuvant Breast Project (Protocol No. 4). XI. Bilateral breast cancer. Cancer 1984; 54:3002–3011. 6498774
13. Carter, C.L., Allen, C., Henson, D.E., Relation of tumor size, lymph node status, and survival in 24,740 breast cancer cases. Cancer 1989; 63:181–187. 2910416
14. Bennett, R.L., Evans, A.J., Kutt, E., et al, Pathological and mammographic prognostic factors for screen detected cancers in a multi-centre randomised, controlled trial of mammographic screening in women from age 40 to 48 years. Breast. 2011;20(6):525–528. 21696957
15. Foulkes, W.D., Grainge, M.J., Rakha, E.A., et al, Tumor size is an unreliable predictor of prognosis in basal-like breast cancers and does not correlate closely with lymph node status. Breast Cancer Res Treat. 2009;117(1):199–204. 18600446
16. O’Dwyer, P.J. Editorial. Axillary dissection in primary breast cancer; the benefits of node clearance warrant reappraisal. Br Med J. 1992; 302:360–361.
17. Blamey, R.W. Clinical aspects of malignant disease. In: Elston C.W., Ellis I.O., eds. Systemic pathology. The Breast. 3rd ed. London: Churchill Livingstone; 1998:501–513.
18. Henson, D.E., Ries, L., Freedman, L.S., et al, Relationship among outcome stage of disease and histologic grade for 22,616 cases of breast cancer. Br Cancer Res Treat 1991; 68:2142–2149. 1913453
19. Kutianawala, M.A., Sayed, M., Stotter, A., et al, Staging the axilla in breast cancer: an audit of lymph-node retrieval in one UK regional centre. Eur J Surg Oncol 1998; 24:280–282. 9724993
20. Steele, R.J.C., Forrest, A.P.M., Gibson, T., The efficacy of lower axillary sampling in obtaining lymph node status in breast cancer: a controlled randomized trial. Br J Surg 1985; 72:368–369. 3888336
21. Dixon, J.M., Dillon, P., Anderson, T.J., et al. Axillary sampling in breast cancer: an assessment of its efficacy. Breast. 1998; 7:206–208.
22. Cabanas, R.M., An approach for the treatment of penile carcinoma. Cancer 1977; 39:456–466. 837331
23. Royal College of Radiologists. Quality assurance guidelines for radiologists, 1990. [NHS BSP Publications no. 15].
24. Royal College of Radiologists. Quality assurance guidelines for radiologists, 1997. [NHS BSP Publications no. 15. January].
25. European Commission. European guidelines for quality assurance in mammography screening, 2nd ed. Luxembourg: Office for Official Publications of the European Communities, 1996.
26. Mansel, R.E., Goyal, A., Newcombe, R.G., ALMANAC Trialists Group, Internal mammary node drainage and its role in sentinel lymph node biopsy: the initial ALMANAC experience. Clin Breast Cancer. 2004;5(4):279–284. 15507173
27. Rosen, P.P., Groshen, S., Saigo, S., et al, Pathological prognostic factors in stage I (TIN0M0) and stage II (TINIM0) breast carcinoma: a study of 644 patients with median follow-up of 18 years. J Clin Oncol 1989; 7:1239–1251. 2549203
28. Parker, C., Rampaul, R.S., Pinder, S.E., et al, E-Cadherin as a prognostic indicator in primary breast cancer. Br J Cancer 2001; 85:1958–1963. 11747340
29. Rakha, E.A., El-Sayed, M.E., Powe, D.G., et al, Invasive lobular carcinoma of the breast: response to hormonal therapy and outcomes. Eur J Cancer. 2008;44(1):73–83. 18035533
30. Elston, C.W., Ellis, I.O., Pathological prognostic factors in breast cancer I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 2002;41(3A):154–161. 12405947
31. Rakha, E.A., El-Sayed, M.E., Lee, A.H., et al, Prognostic significance of Nottingham histologic grade in invasive breast carcinoma. J Clin Oncol. 2008;26(19):3153–3158. 18490649
32. Rakha, E.A., El-Sayed, M.E., Menon, S., et al, Histologic grading is an independent prognostic factor in invasive lobular carcinoma of the breast. Breast Cancer Res Treat. 2008;111(1):121–127. 17929165
33. Pinder, S.E., Ellis, I.O., Galea, M., et al, Pathological prognostic factors in breast cancer III. Vascular invasion: relationship with recurrence and survival in a large study with long-term follow-up. Histopathology. 1994;24(1):41–47. 8144141
34. Truong, P.T., Yong, C.M., Abnousi, F., et al, Lymphovascular invasion is associated with reduced locoregional control and survival in women with node-negative breast cancer treated with mastectomy and systemic therapy. J Am Coll Surg. 2005;200(6):912–921. 15922205
35. Haybittle, J.L., Blamey, R.W., Elston, C.W., et al, A prognostic index in primary breast cancer. Br J Cancer 1982; 45:361–366. 7073932
36. Ellis, I.O., Bell, J., Todd, J., et al, Evaluation of immunoreactivity with monoclonal antibody NCRC-II in breast carcinoma. Br J Cancer 1987; 56:295–299. 3663477
37. Lee, A.H., Pinder, S.E., Macmillan, R.D., et al, Prognostic value of lymphovascular invasion in women with lymph node negative invasive breast carcinoma. Eur J Cancer. 2006;42(3):357–362. 16377180
38. Gee, J.M., Eloranta, J.J., Ibbitt, J.C., et al, Overexpression of TFAP2C in invasive breast cancer correlates with a poorer response to anti-hormone therapy and reduced patient survival. J Pathol. 2009;217(1):32–41. 18825690
39. Sarwar, N., Kim, J.S., Jiang, J., et al, Phosphorylation of ERalpha at serine 118 in primary breast cancer and in tamoxifen-resistant tumours is indicative of a complex role for ERalpha phosphorylation in breast cancer progression. Endocr Relat Cancer. 2006;13(3):851–861. 16954434
40. Gee, J.M., Shaw, V.E., Hiscox, S.E., et al, Deciphering antihormone-induced compensatory mechanisms in breast cancer and their therapeutic implications. Endocr Relat Cancer. 2006;13(Suppl. 1):S77–S88. 17259561
41. Dowle, C.S., Owainati, A., Robins, A., et al, The prognostic significance of the DNA content of human breast cancer. Br J Surg 1987; 74:133–136. 3815031
42. Putti, T.C., El-Rehim, D.M., Rakha, E.A., et al, Estrogen receptor-negative breast carcinomas: a review of morphology and immunophenotypical analysis. Mod Pathol. 2005;18(1):26–35. 15332092
43. Dowsett, M., Allred, C., Knox, J., et al, Relationship between quantitative estrogen and progesterone receptor expression and human epidermal growth factor receptor 2 (HER-2) status with recurrence in the Arimidex, Tamoxifen, Alone or in Combination trial. J Clin Oncol. 2008;26(7):1059–1065. 18227529
44. Kumar, R.R., Meenakshi, A., Sivakumar, N., Enzyme immunoassay of human epidermal growth factor (hEGFR). Hum Antibodies 2001; 10:143–147. 11847425
45. Eberhard, D.A., Huntzicker, E., et al. Epidermal Growth Factor receptor immunohistochemistry: assay selection and amplification to breast cancers. ASCO. 2002. [no. 1791 (Abstr.)].
46. Klijn, J.G.M., Berns, P.M.J., Schmitz, P.I., et al, The clinical significance of epidermal growth factor receptor in human breast cancer: a review of 5232 patients. Endocr Rev 1992; 13:3–17. 1313356
47. Tsutsui, S., Ohno, S., Murakami, S., et al, Prognostic value of epidermal growth factor and its relationship to the ER status of 1029 patients with breast cancer. Breast Cancer Res Treat 2002; 71:67–75. 11859875
48. Rampaul, R.S., Pinder, S.E., Robertson, J.F., et al, EGFR expression in operable breast cancer: is it of prognostic significance? Clin Cancer Res. 2004;10(7):2578. 15073139
49. Ferrero, J.M., Ramaioli, A., Largillier, R., et al, Epidermal growth factor receptor expression in 780 breast cancer patients: a reappraisal of the prognostic value based on an eight-year median follow-up. Ann Oncol. 2001;12(6):835–841. 11484961
50. Nicholson, R.I., McCelland, R.A., Gee, J.M.W., et al, Epidermal growth factor receptor expression in breast cancer. Association with response to endocrine therapy. Breast Cancer Res Treat 1994; 29:117–125. 7912565
51. Knoop, A., Bentzen, S.M., Nielsen, M.M., et al, Value of epidermal growth factor receptor, HER-2, p53 and steroid receptors in predicting the efficacy of Tamoxifen in high risk postmenopausal breast cancer patients. J Clin Oncol 2001; 19:3376–3384. 11454885
52. Wolff, A.C., Hammond, M.E., Schwartz, J.N., et al, American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol 2007; 25:118–145. 17159189
53. Ratcliffe, N., Wells, W., Wheeler, K., et al, The combination of in situ hybridization and immunohistochemical analysis: an evaluation of Her2/neu expression in paraffin-embedded breast carcinomas and adjacent normal-appearing breast epithelium. Mod Pathol 1997; 10:1247–1252. 9436971
54. Busmanis, I., Feleppa, F., Jones, A., et al, Analysis of cerbB2 expression using a panel of 6 commercially available antibodies. Pathology 1994; 26:261–267. 7991280
55. Carlson, R.W., Moench, S.J., Hammond, M.E., et al, HER2 testing in breast cancer: NCCN Task Force report and recommendations. J Natl Compr Canc Netw. 2006;4(Suppl. 3):S1–24. 16813731
56. Bartlett, J.M., Going, J.J., Mallon, E.A., et al, Evaluating HER2 amplification and overexpression in breast cancer. J Pathol 2001; 195:422–428. 11745673
57. Press, M.F., Slamon, D.J., Flom, K.J., et al, Evaluation of HER-2/neu gene amplification and overexpression: comparison of frequently used assay methods in a molecularly characterized cohort of breast cancer specimens. J Clin Oncol 2002; 20:3095–3105. 12118023
58. Walker, R.A., Bartlett, J.M., Dowsett, M., et al, HER2 testing in the UK: further update to recommendations. J Clin Pathol. 2008;61(7):818–824. 18381380
59. Dowsett, M., Bartlett, J., Ellis, I.O., et al, Correlation between immunohistochemistry (HercepTest) and fluorescence in situ hybridization (FISH) for HER-2 in 426 breast carcinomas from 37 centres. J Pathol 2003; 199:418–423. 12635131
60. Slamon, D.J., Clark, G.M., Wong, S.G., et al, Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177–182. 3798106
61. Rilke, F., Colnaghi, M.I., Cascinelli, N., et al, Prognostic significance of HER-2/neu expression in breast cancer and its relationship to other prognostic factors. Int J Cancer. 1991;49(1):44–49. 1678734
62. Viani, G.A., Afonso, S.L., Stefano, E.J., et al, Adjuvant trastuzumab in the treatment of her-2-positive early breast cancer: a meta-analysis of published randomized trials. BMC Cancer 2007; 7:153. 17686164
63. Carter, P., Presta, L., Gorman, C.M., et al, Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci U S A 1992; 89:4285–4289. 1350088
64. Disis, M.L., Knutson, K.L., Schiffman, K., et al, Pre-existent immunity to the HER-2/neu oncogenic protein in patients with HER-2/neu overexpressing breast and ovarian cancer. Breast Cancer Res Treat 2000; 62:245–252. 11072789
65. Ward, R.L., Hawkins, N.J., Coomber, D., et al, Antibody immunity to the HER-2/neu oncogenic protein in patients with colorectal cancer. Hum Immunol 1999; 60:510–515. 10408800
66. Mirza, A.N., Mirza, N.Q., Vlastos, G., et al, Prognostic factors in node-negative breast cancer: a review of studies with sample size more than 200 and follow-up more than 5 years. Ann Surg. 2002;235(1):10–26. 11753038
67. Bernards, R., Destree, A., McKenzie, S., et al, Effective tumor immunotherapy directed against an oncogene-encoded product using a vaccinia virus vector. Proc Natl Acad Sci U S A 1987; 84:6854–6858. 3477812
68. Amar, S., McCullough, A.E., Tan, W., et al, Prognosis and outcome of small (≤ 1 cm), node-negative breast cancer on the basis of hormonal and HER-2 status. Oncologist. 2010;15(10):1043–1049. 20930097
69. Chia, S., Norris, B., Speers, C., et al, Human epidermal growth factor receptor 2 overexpression as a prognostic factor in a large tissue microarray series of node-negative breast cancers. J Clin Oncol 2008; 26:5697–5704. 19001334
70. Kraus, M.H., Popescu, N.C., Amsbaugh, S.C., et al, Overexpression of the EGF receptor-related proto-oncogene erbB-2 in human mammary tumor cell lines by different molecular mechanisms. EMBO J 1987; 6:605–610. 3034598
71. Ring, C.J., Blouin, P., Martin, L.A., et al, Use of transcriptional regulatory elements of the MUC1 and ERBB2 genes to drive tumour-selective expression of a prodrug activating enzyme. Gene Ther 1997; 4:1045–1052. 9415310
72. Yu, D.H., Hung, M.C., Expression of activated rat neu oncogene is sufficient to induce experimental metastasis in 3T3 cells. Oncogene 1991; 6:1991–1996. 1682865
73. Bria, E., Cuppone, F., Fornier, M., et al, Cardiotoxicity and incidence of brain metastases after adjuvant trastuzumab for early breast cancer: the dark side of the moon? A meta-analysis of the randomized trials. Breast Cancer Res Treat. 2008;109(2):231–239. 17638068
74. Chang, H., Riese, D.J., 2nd., Gilbert, W., et al, Ligands for ErbB-family receptors encoded by a neuregulin-like gene. Nature 1997; 387:509–512. 9168114
75. Juhl, H., Downing, S.G., Wellstein, A., et al, HER-2/neu is rate-limiting for ovarian cancer growth. Conditional depletion of HER-2/neu by ribozyme targeting. J Biol Chem 1997; 272:29482–29486. 9368008
76. Deshane, J., Siegal, G.P., Wang, M., et al, Transductional efficacy and safety of an intraperitoneally delivered adenovirus encoding an anti-erbB-2 intracellular single-chain antibody for ovarian cancer gene therapy. Gynecol Oncol 1997; 64:378–385. 9062138
77. Schmidt, M., Hynes, N.E., Groner, B., et al, A bivalent single-chain antibody-toxin specific for ErbB-2 and the EGF receptor. Int J Cancer 1996; 65:538–546. 8621240
78. Park, J.W., Hong, K., Carter, P., et al, Development of anti-p185HER2 immunoliposomes for cancer therapy. Proc Natl Acad Sci U S A 1995; 92:1327–1331. 7877976
79. Chen, S.Y., Yang, A.G., Chen, J.D., et al, Potent antitumour activity of a new class of tumour-specific killer cells. Nature 1997; 385:78–80. 8985250
80. Newby, J.C., Johnston, S.R.D., Smith, I., et al, Expression of epidermal growth factor and C-erb-2 during the development of tamoxifen resistance in human breast cancer. Clin Cancer Res 1997; 3:1643–1651. 9815855
81. Nicholson, R.I., McCelland, R.A., Finlay, P., et al, Relationship between EGF-R, C-erb-2 protein expression and Ki67 immunostaining in breast cancer and hormone sensitivity. Eur J Cancer 1993; 29A:1018–1023. 8098946
82. Giai, M., Roagna, R., Ponzone, R., et al, Prognostic and predictive relevance of C-erb-2 and ras expression in node positive and negative breast cancer. Anticancer Res 1994; 14:1441–1450. 7915096
83. Archer, S.G., Eliopoulos, S.A., Spandidos, D., et al, Expression of ras p21, p53 and C-erb-2 in advanvced breast cancer and response to first line hormonal therapy. Br J Cancer 1995; 72:1259–1266. 7577479
84. Pegram, M.D., Konecny, G.E., O’Callaghan, C., et al, Rational combinations of trastuzumab with chemotherapeutic drugs used in the treatment of breast cancer. J Natl Cancer Inst. 2004;96(10):739–749. 15150302
85. Muss, H., Berry, D., Thor, A., et al. Lack of interaction of tamoxifen (T) use and ErbB-2/HER-2/neu (H) expression in CALGB 8541: a randomized adjuvant trial of three different doses of cyclophosphamide, doxorubicin and fluorouracil (CAF) in node-positive primary breast cancer (BC). Proc Am Soc Clin Oncol. 1999; 18:68a.
86. Paik, S., Bryant, J., Park, C., et al, ErbB-2 and response to doxorubicin in patients with axillary lymph node positive, hormone receptor-negative breast cancer. J Natl Cancer Inst 1998; 90:1361–1370. 9747867
87. Elledge, R.M., Green, S., Ciocca, D., et al, HER-2 expression and response to tamoxifen in estrogen receptor-positive breast cancer: a Southwest Oncology Group Study. Clin Cancer Res. 1998;4(1):7–12. 9516946
88. Ravin, P.M., Green, S., Albain, V., et al. Initial report of the SWOG biological correlative study of CerbB-2 expression as a predictor of outcome in a trial comparing adjuvant CAF with tamoxifen (T) alone. Proc Am Soc Clin Oncol. 1998; 17:97a.
89. Rakha, E.A., Reis-Filho, J.S., Ellis, I.O., Basal-like breast cancer: a critical review. J Clin Oncol. 2008;26(15):2568–2581. 18487574
90. Johannsson, O.T., Idvall, I., Anderson, C., et al, Tumour biological features of BRCA1-induced breast and ovarian cancer. Eur J Cancer 1997; 33:362–371. 9155518
91. Chappuis, P.O., Nethercot, V., Foulkes, W.D., Clinico-pathological characteristics of BRCA1- and BRCA2-related breast cancer. Semin Surg Oncol 2000; 18:287–295. 10805950
92. Marcus, J.N., Watson, P., Page, D.L., Hereditary breast cancer. Pathobiology, prognosis and BRCA 1 and BRCA 2 linkage. Cancer 1996; 77:697–709. 8616762
93. Marcus, J.N., Page, D.L., Watson, P., et al. BRCA 1 and BRCA 2 hereditary breast cancer phenotypes. Cancer. 1997; 80:543.
94. Breast Cancer Linkage Consortium, Pathology of familial breast cancer: differences between breast cancer in carriers of BRCA 1 or BRCA 2 mutational and sporadic cases. Lancet 1997; 349:1505. 9167459
95. Robson, M., Gilewski, T., Haas, B., et al, BRCA-associated breast cancer in young women. J Clin Oncol 1998; 16:1642–1649. 9586873
96. Karp, S.E., Tonin, P.N., Bégin, L.R., et al, Influence of BRCA 1 mutations on nuclear grade and oestrogen receptor status of breast carcinoma in Ashkenazi Jewish women. Cancer 1997; 80:435–441. 9241077
97. Armes, J.E., Egan, A.J.M., Southey, M.C., et al, The histologic phenotypes of breast carcinoma occurring before age 40 in women with and without BRCA 1 or BRCA 2 germline mutations. Cancer 1998; 83:2335–2345. 9840533
98. Agnarsson, B.A., Jonasson, J.G., Björnsdottir, I.B., et al, Inherited BRCA 2 mutation associated with high grade breast cancer. Breast Cancer Res Treat 1998; 47:121–127. 9497100
99. Marcus, J.N., Watson, P., Page, D.L., et al, BRCA 2 hereditary breast cancer phenotype. Breast Cancer Res Treat 1997; 44:275–277. 9266108
100. Baak, J.P., Colpaert, C.G., van Diest, P.J., et al, Multivariate prognostic evaluation of the mitotic activity index and fibrotic focus in node-negative invasive breast cancers. Eur J Cancer. 2005;41(14):2093–2101. 16153819
101. Baak, J.P.A., Vooiji, G.P., Brugal, G. Nuclear image cytometry: quantitation of chromatin pattern, steroid receptor content and Ki-67. In: Baale J.P.A., ed. Manual of quantitative pathology in cancer diagnosis and prognosis. Heidelberg: Springer-Verlag; 1991:232–243.
102. Palcic, B., Garner, D.M., MacAulay, C.E., Image cytometry and chemoprevention in cervical cancer. J Cell Biochem. 1995;23(Suppl.):43–54. 8747377
103. Nagahata, T., Hirano, A., Utada, Y., et al, Correlation of allelic losses and clinicopathologic factors in 504 primary breast cancers. Breast Cancer 2002; 9:208–215. 12185331
104. Hermsen, M.A.J.A., Baak, J.P.A., Weiss, J., et al, Genetic analysis of 513 lymph node negative breast carcinomas by CGH and relation to clinical, pathologic, morphometric and DNA cytometric prognostic factors. J Pathol 1998; 186:356–362. 10209483
105. Janssen, E.A.M., Baak, J.P.A., Guervos, M.A., et al, In lymph node-negative breast cancer, specific chromosomal aberrations are strongly associated with high mitotic activity and predict outcome more accurately than grade, tumour diameter, and oestrogen receptor. J Pathol. 2003;201(4):555–561. 14648658
106. Hyman, E., Kauraniemi, P., Hautaniemi, S., et al, Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res 2002; 62:6240–6245. 12414653
107. Hu, Z., Fan, C., Oh, D.S., et al, The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics 2006; 7:96. 16643655
108. Hedenfalk, I., Duggan, D., Chen, Y., et al, Gene expression profiles in hereditary breast cancer. N Eng J Med 2001; 344:539–548. 11207349
109. van’t Veer, L.J., Dai, H., van de Vijer, M.J., et al, Expression profiling predicts outcomes in breast cancer. Breast Cancer Res 2002; 5:57–58. 12559048
110. Gruvberger, S., Ringner, M., Chen, Y., et al, Oestrogen receptor status in breast cancer is associated with remarkably distinct gene expression profiles. Cancer Res 2001; 61:5979–5984. 11507038
111. West, M., Blanchette, C., Dressman, H., et al, Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci U S A 2001; 98:11462–11467. 11562467
112. Ahr, A., Kam, T., Solbach, S., et al, Identification of high risk breast cancer by gene expression profiling. Lancet 2002; 359:131–132. 11809257
113. van de Vijver, M.J., He, Y.D., van’t Veer, L.J., A gene expression signature as a predictor of survival in breast cancer. N Engl J Med 2002; 347:1999–2009. 12490681
114. Leong, A.S.-Y., Zhuang, Z., The changing role of pathology in breast cancer diagnosis and treatment. Pathobiology. 2011;78(2):99–114. 21677473
115. Bertucci, F., Viens, P., Hingamp, P., et al, Breast cancer revisited using DNA array-based gene expression profiling. Int J Cancer. 2003;103(5):565–571. 12494462
116. Perou, C.M., Sorlie, T., Eisen, M.B., et al, Molecular portraits of human breast tumours. Nature 2000; 406:747–752. 10963602
117. Habel, L.A., Shak, S., Jacobs, M.K., et al, A population-based study of tumor gene expression and risk of breast cancer death among lymph node-negative patients. Breast Cancer Res 2006; 8:R25. 16737553
118. Chang, H.Y., Nuyten, D.S., Sneddon, J.B., et al, Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A 2005; 102:3738–3743. 15701700
119. Chi, J.T., Wang, Z., Nuyten, D.S., et al, Gene expression programmes in response to hypoxia: cell type specificity and prognostic significance in human cancers. PLoS Med 2006; 3:e47. 16417408
120. McCelland, C.M., Gullick, W.J., Identification of surrogate markers for determining drug activity using proteomics. Biochem Soc Trans. 2003;31(6):1488–1490. 14641096
121. Chevallier, B., Mossen, V., Dauce, J.P., et al, A prognostic score in histological node negative breast cancer. Br J Cancer 1990; 61:436–440. 2328212
122. Brown, J.M., Benson, E.A., Jones, M. Confirmation of a long term prognostic index in breast cancer. Breast. 1993; 2:144–147.
123. Balslev, I., Axesson, C.K., Zedelev, K., et al, The Nottingham Prognostic Index applied to 9,149 patients from the studies of the Danish Breast Cancer Cooperative Group (DBCG). Br Cancer Res Treat 1994; 32:281–290. 7865856
124. Ravdin, P.M., Siminoff, L.A., Davis, G.J., et al, Computer programme to assist in making decisions about adjuvant therapy for women with early breast cancer. J Clin Oncol 2001; 19:980–991. 11181660
125. Goldhirsch, A., Glick, J.H., Gelber, R.D., et al, Meeting highlights: international expert consensus on the primary therapy of early breast cancer 2005. Ann Oncol 2005; 16:1569–1583. 16148022
126. Wishart, G.C., Azzato, E.M., Greenberg, D.C., et al, PREDICT: a new UK prognostic model that predicts survival following surgery for invasive breast cancer. Breast Cancer Res. 2010;12(1):R1. 20053270
127. Robertson, J.F., Ellis, I.O., Pearson, D., et al, Biological factors of prognostic significance in locally advanced breast cancer. Breast Cancer Res Treat. 1994;29(3):259–264. 8049459
128. Robertson, J.F., Dixon, A.R., Nicholson, R.I., et al, Confirmation of a prognostic index for patients with metastatic breast cancer treated by endocrine therapy. Breast Cancer Res Treat. 1992;22(3):221–227. 1391988
129. Ellis IO, et al. Improvement of the Nottingham Prognostic Index (NPI +). Cancer Res (In Press).