Molecular Techniques
At the conclusion of this chapter, the reader should be able to:
• Describe the composition of DNA and RNA.
• Compare the functions of DNA and various forms of RNA.
• Describe the polymerase chain reaction (PCR) amplification technique.
• Compare various PCR modifications.
• Identify and briefly describe other amplification techniques.
• Describe the gold standard of genetic analysis.
• Compare DNA sequencing and branched DNA protocols.
• Identify and compare three hybridization techniques.
• Explain how microarrays are applied to immunologic testing.
• Discuss the general concept of nucleic acid blotting.
• Compare the characteristics and clinical applications of Southern, Northern, and Western blotting techniques.
• Analyze a case study related to immunoassay.
• Correctly answer case study related multiple choice questions.
• Be prepared to participate in a discussion of critical thinking questions.
Characteristics of Nucleic Acids
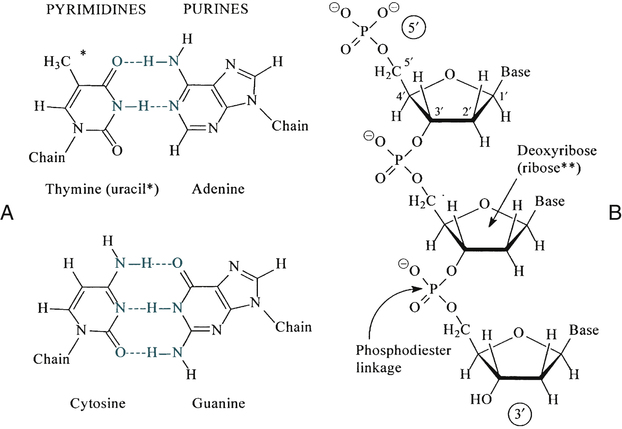
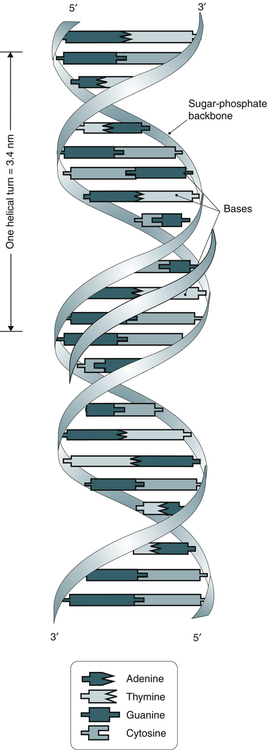
DNA and RNA are polymers made up of repeating nucleotides or bases that are linked together (Fig. 14-1). DNA and RNA have the same two purine bases, adenine (A) and guanine (G), but the pyrimidine bases differ. DNA has cytosine (C) and thymine (T); RNA substitutes uracil (U) for T. DNA is predominantly a double-stranded molecule with specific base pairs linked together (Fig. 14-2). Nucleotides are bonded together and two strands are twisted into an alpha helix (Fig. 14-3).
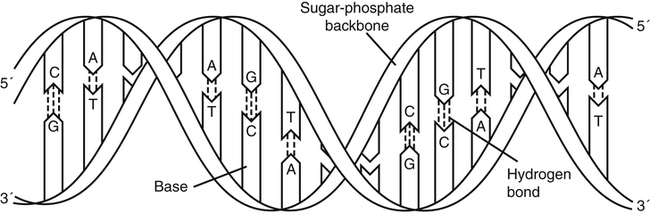
The DNA molecule is a double helix that consists of two sugar-phosphate backbones with four bases—cytosine (C), guanine (G), adenine (A), and thymine (T)—attached. C and G residues and A and T residues on opposite strands pair through hydrogen bonding. (From LeGrys V, Leinbach SS, Silverman L: Clinical applications of DNA probes in the diagnosis of genetic diseases, Crit Rev Clin Lab Sci 25:255, 1987.)
How Does DNA Replicate?
DNA is a very stable molecule and replication is straightforward. The process of replication (Fig. 14-4) involves one strand of the molecule acting as a template for the creation of a complementary strand. As a result of this process, two identical daughter molecules are produced. In the laboratory, the hydrogen bonds that hold the strands of the double helix can be broken apart or denatured. If complementary strands of DNA are denatured in the laboratory, they can spontaneously rejoin, or anneal. The process of denaturation and annealing (see later discussion) can be used effectively in molecular testing.
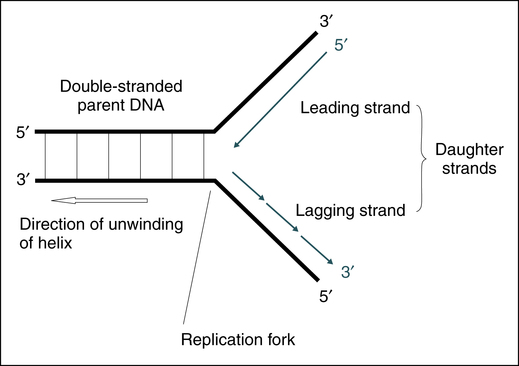
Double-stranded DNA is separated at the replication fork. The leading strand is synthesized continuously, whereas the lagging strand is synthesized discontinuously but joined later by DNA ligase. (From Burtis CA, Ashwood ER, Bruns DB: Tietz fundamentals of clinical chemistry, ed 6, St Louis, 2008, Saunders.)
Forms of RNA
• mRNA—translates DNA coding into functional proteins (Fig. 14-5)
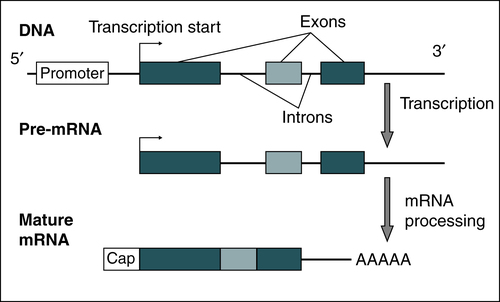
A gene that encodes for a protein contains a promoter region with a variable number of introns and exons. Transcription commences at the transcription start site. Pre-mRNA is processed by capping, polyadenylation, and intron splicing and becomes a mature mRNA. (From Burtis CA, Ashwood ER, Bruns DB: Tietz fundamentals of clinical chemistry, ed 6, St Louis, 2008, Saunders.)
• tRNA—transports various amino acids to manufacture proteins
Amplification Techniques in Molecular Biology
Polymerase Chain Reaction
The polymerase chain reaction (PCR) is an in vitro method that amplifies low levels of specific DNA sequences in a sample to higher levels suitable for further analysis (Fig. 14-6, A). To use this technology, the target sequence to be amplified must be known. Typically, a target sequence ranges from 100 to 1000 base pairs (bp) in length. Two short DNA primers, typically 16 to 20 bp, are used. The oligonucleotides (small portions of a single DNA strand) act as a template for the new DNA. These primer sequences are complementary to the 3′ ends of the sequence to be amplified.
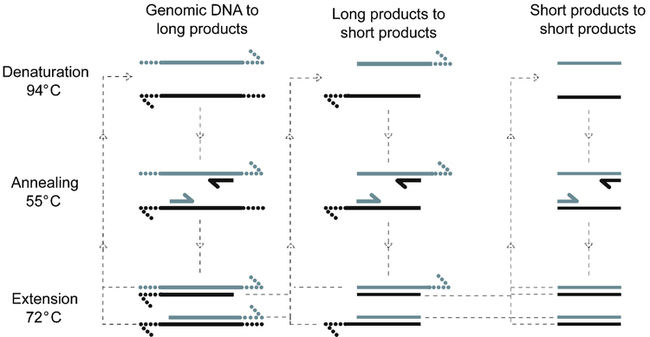
Repetitive cycles of denaturation, annealing, and extension are paced by temperature cycling of the reaction. Two primers indicated as short segments anneal to opposite template strands (long line) to define the region to be amplified. Extension occurs from the 3′ ends (half-arrowheads). In each cycle, genomic DNA is denatured and annealed to primers that extend in opposite directions across the same region, producing long products of undefined length. Long products generated by extension of one of the primers anneal to the other primer during the next cycle, producing short products of defined length. Any short products present also produce more short products. After n cycles, up to 2n new copies of the amplified region are present—n long products and (2nn − n) short products plus one original genomic copy. A similar approach can be used to amplify RNA targets by initial reverse transcription of the RNA template to produce the DNA template. (From Burtis CA, Ashwood ER, Bruns DB: Tietz fundamentals of clinical chemistry, ed 6, St Louis, 2008, Saunders.)
This enzymatic process is carried out in cycles. Each repeated cycle consists of the following:
• DNA denaturation. Separation of the double DNA strands into two single strands through the use of heat.
• Primer annealing. Recombination of the oligonucleotide primers with the single-stranded original DNA.
• Extension of primed DNA sequence. The enzyme DNA polymerase synthesizes new complementary strands by the extension of primers.
Three important applications of PCR are as follows:
The PCR technique has undergone modifications (see Fig. 14-6, B). One uses nested primers in a two-step amplification process. First, a broad region of the DNA surrounding the sequence of interest is amplified, followed by another round of amplification to amplify the specific gene sequence to be studied. Another PCR modification successfully differentiates alleles of the same gene.
Analysis of Amplification Products
Other Techniques
DNA Sequencing
1. The first step in sequencing a target is usually to amplify it by cloning or in vitro amplification, usually PCR. Once the amplified DNA is purified from the clinical specimen (the target DNA), it is heat-denatured to separate the double-stranded DNA (dsDNA) into single strands (ssDNA).
2. The second step involves adding primers to the ssDNA. Primers are short synthetic segments of ssDNA that contain a nucleotide sequence complementary to a short strand of target DNA. The patient’s DNA serves as a template to copy. DNA polymerase catalyzes the addition of the appropriate nucleotides to the preexisting primer. DNA synthesis is terminated when the deoxynucleotide is incorporated into a growing DNA chain.
Hybridization Techniques
Blotting Protocols
1. Electrophoretic separation of the patient’s nucleic acid
2. Transfer of nucleic acid fragments to a solid support (e.g., nitrocellulose)
3. Hybridization with a labeled probe of known nucleic acid sequence
4. Autoradiographic or colorimetric detection of the bands created by the probe–nuclei acid hybrid
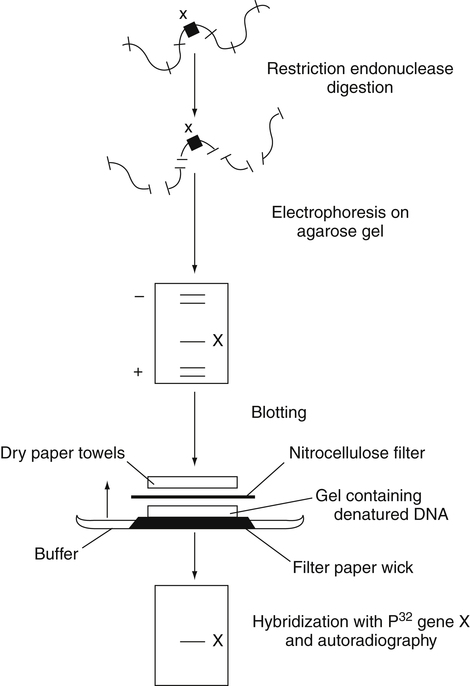
Southern Blot
Specimen DNA is denatured and treated with restriction enzymes to create DNA fragments; then the ssDNA fragments are separated by electrophoresis (Figure 14-7). The electrophoretically separated fragments are then blotted to a nitrocellulose membrane, retaining their electrophoretic position and hybridized with radiolabeled single-stranded DNA fragments with sequences complementary to those being sought. The resulting dsDNA bearing the radiolabel, if present, is then detected by radiography.
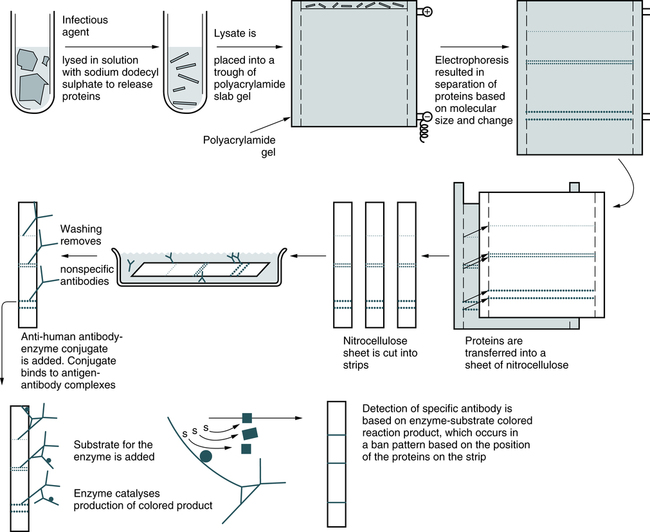
Western Blot
Compared with the Southern blot technique, which separates and identifies RNA fragments and proteins, and the Northern blot technique, which concentrates on isolating mRNA, in the Western blot technique proteins are separated electrophoretically, transferred to membranes, and identified through the use of labeled antibodies specific for the protein of interest (Fig. 14-8).
Microarrays
Microarray (DNA chip) technology has helped accelerate genetic analysis, just as microprocessors accelerated computation (Fig. 14-9). Microarrays are basically the product of bonding or direct synthesis of numerous specific DNA probes on a stationary, often silicon-based support. The chip may be tailored to particular disease processes. The technique is easily performed and readily automated.
Microarrays are miniature gene fragments attached to glass chips. These chips are used to examine the gene activity of thousands or tens of thousands of gene fragments and to identify genetic mutations using a hybridization reaction between the sequences on the microarray and a fluorescent sample. After hybridization, the chips are scanned with high-speed fluorescent detectors and the intensity of each spot is quantitated (Fig. 14-10).
The identity and amount of each sequence are revealed by the location and intensity of fluorescence displayed by each spot. Computers are used to analyze the data (Fig. 14-11).
Next Generation Sequencing Technology
With NGS technology, the process begins with template preparation by shearing DNA (or cDNA) to create fragment libraries. Adaptor sequences are added to these fragments and serve as primers for amplification usually by emulsion PCR or bridge PCR methods. The resulting amplified signal beads or clusters are analyzed using a variety of platform-specific chemical analyses but all are based on the addition of labeled nucleotides. Digital images are captured and analyzed to determine the sequence of the target DNA. The impact of NGS on cancer treatment is presented in Chapter 33, Tumor Immunology.
Chapter Highlights
• The polymerase chain reaction (PCR) is an in vitro method that amplifies low levels of specific DNA sequences in a sample to higher levels suitable for further analysis.
• PCR, an enzymatic process, is carried out in cycles. Each repeated cycle consists of DNA denaturation, primer annealing, and extension of the primed DNA sequence. Each cycle theoretically doubles the amount of specific DNA sequence present and results in an exponential accumulation of the DNA fragment being amplified (amplicons).
• PCR analysis can lead to the detection of gene mutations, identification of viral DNA associated with specific cancers, and detection of genetic mutations.
• Adaptations of the PCR technique include nested primers. Modifications include RT-PCR, multiplex PCR, and real-time PCR.
• Conventional analysis uses agarose gel electrophoresis after ethidium bromide staining.
• Probe-based DNA detection systems provide sequence specificity and lower detection limits.
• Selection of technique is based on sensitivity and specificity profiles, cost, turnaround time, and local experience.
• Probe hybridization assays involve the complementary pairing of a probe with DNA or RNA from the patient’s specimen; these include liquid-phase, dot blot, and reverse dot blot assays.
• The Southern blot technique can determine single-base mutations (e.g., sickle cell anemia, hemophilia A).
• The Northern blot technique can be used for the detection of specific mRNA. This procedure is not routinely used in clinical molecular diagnostics.
• In the Western blot technique, proteins are separated electrophoretically, transferred to membranes, and identified through labeled antibodies. It is used to detect antibodies to specific epitopes of antigen subspecies and confirm the specificity of antibodies detected by ELISA screening.
• Microarrays (DNA chips) are the product of bonding or synthesis of specific DNA probes on a stationary support. These chips are used to examine gene activity and identify genetic mutations in malignancies, test for genetic disease test, and detect virally resistant mutations.