Molecular biology
Selected techniques used in the analysis of DNA
Fluorescence in situ hybridisation (FISH)
FISH describes the hybridisation of specific DNA or RNA sequences in situ to cellular targets attached to microscope slides. The most popular probes are chromosome-specific DNA sequences which generate a brilliant signal in both metaphase and interphase nuclei. The technique is particularly useful in the demonstration of chromosomal monosomies or trisomies but chromosome translocations (Fig 50.1), deletions and amplification of specific genes can also be detected. The results of FISH may be further improved by image processing.
Microarrays/gene profiling
Microarray technology allows the simultaneous profiling of tens of thousands of genes thus painting a molecular portrait of a tumour cell. Several methods are available for analysis of a large number of RNA transcripts. These include complementary DNA microarrays, oligonucleotide microarrays and serial analysis of gene expression (SAGE). The most commonly used ‘platforms’ are the two microarray technologies in which each experiment reveals the expression levels of over 20 000 genes. cDNA fragments and oligonucleotides can be spotted onto glass slides. The DNA arrayed on the slide is generally referred to as the ‘probe’ and the cDNA or cRNA derived from the sample is referred to as the ‘target’. The complex gene expression data generated requires powerful statistical analysis. Microarrays are enhancing our understanding of haematological malignancy (see below and Fig 50.2).
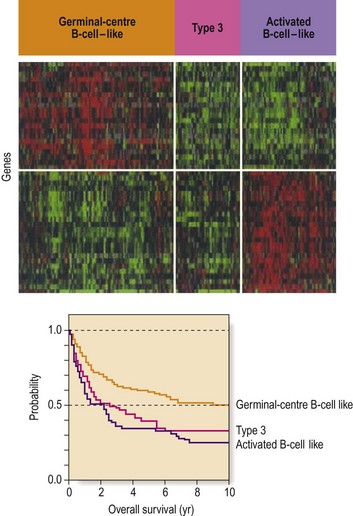
Fig 50.2 Subgroups of diffuse large B-cell non-Hodgkin’s lymphoma (DLBCL) according to gene expression profiles.
In the top panel each column represents a single DLBCL and each row a single gene. Red areas indicate increased expression and green areas reduced expression. Survival after chemotherapy is different in the three groups. (Reprinted with permission from Margalit O et al 2005 Micro-array based gene expression profiling of hematologic malignancies: basic concepts and clinical applications. Blood Reviews 19: 226.)
Next-generation sequencing
Massively parallel sequencing (also termed next- or second-generation sequencing) results in the simultaneous generation of millions of short DNA sequences. In studies of haematological malignancy, it is important to sequence both the tumour cells and normal tissue (e.g. skin) from the patient to identify relevant acquired (somatic) mutations. In whole genomic sequencing (Fig 50.3), the object is to sequence the entire genome. More specific next-generation techniques include exome and transcriptome sequencing.
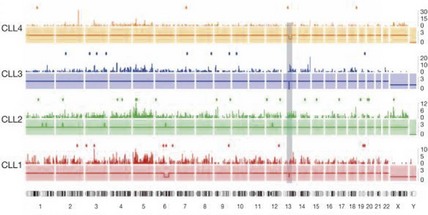
Fig 50.3 Whole genome sequencing for detection of mutations in chronic lymphocytic leukaemia.
For each tumour genome, copy number (solid lines), density of mutations per 5-Mb window (bars) and protein-coding mutations (dots) are shown. The shaded rectangle indicates the location of the 13q14 deletion present in three of the four cases. Chromosome numbers are listed below the four profiles. (Reprinted by permission from Macmillan Publishers Ltd: Puente XS et al 2011, Whole genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 475: 101–105.)
Application of molecular biology in haematology
Haematological malignancy
Minimal residual disease
Traditional definitions of remission in leukaemia have relied on crude morphological criteria. Many patients in remission subsequently relapse, implying the existence of occult neoplastic cells undetectable by normal morphological or cytogenetic methods – so-called minimal residual disease (MRD) (Fig 50.4). Reliable detection of MRD potentially allows improved management with escalation of therapy for patients with persistent disease and the avoidance of excessive treatment in patients showing a good response to previous intervention. Detection of MRD relies upon the presence of disease markers that can be targeted (e.g. PML-RAR α in acute promyelocytic leukaemia). In childhood and adolescent ALL the tandem application of flow cytometry and PCR can be used to study MRD in almost all patients and this information is being employed in clinical trials. In CML quantitative PCR assay of BCR-ABL transcripts is routinely used to direct management. Very low levels of BCR-ABL mRNA predict a good clinical outcome.
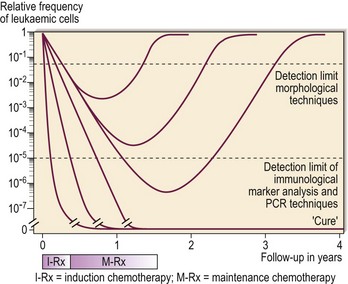
Fig 50.4 The detection of minimal residual disease.
The greater sensitivity of PCR and immunological marker analysis compared with traditional morphological techniques in the detection of residual leukaemic cells can be seen. (Reproduced with permission of JJM van Dongen, Department of Immunology, Erasmus University, Rotterdam, and Medicultura International B.V.)