Chapter 91 Infection and Host Response
Historical Perspectives
Traditionally, the immune system has been divided into innate and adaptive components. Clonal expansion of lymphocytes in response to infection is absolutely critical to the development of the immune response. However, it takes 3 to 5 days for clonal expansion to produce sufficient numbers of “effector” cells. Clearly, this is more than enough time for a pathogen to damage the host. The innate immune system is fundamental in eliminating the infection, and if not, then controlling it, until the adaptive immune responses eliminate it. If the innate and adaptive immune responses are “adequate,” the infection remains localized. If not, then the systemic response to infection, or “sepsis” results (see Chapters 90 and 103). What has become increasingly clear is that the adaptive and innate immune systems can each affect the functioning of the other.1
In the past 25 years, new concepts that are fundamental to the care of critically ill patient have arisen directly from our understanding of the host’s response to infection. (1) Inflammatory reactions first characterized as a response to infection are the foundation of a number of other pathogenic mechanisms, such as ischemia/reperfusion injury, direct trauma, drug-induced injury, inhalational injury, and multiple-system organ dysfunction. (2) The response to infection may lead to further injury of the host. Alternatively, an exuberant antiinflammatory response may likewise be deleterious, resulting in immune suppression and fibrosis (see Chapter 104). (3) There is a direct interaction between the neuroendocrine axis and inflammation (see Chapter 102). (4) There is a fundamental interrelationship among endothelium, inflammation, and coagulation, and efforts to intervene in one will likely have effects in others (see Chapter 101).
Innate Immune Versus Adaptive Immune Response
The innate immune system is phylogenetically ancient. Innate immune recognition is genetically predetermined (i.e., by germline-encoded receptors). Thus these receptors have evolved by natural selection, and have defined specificities for infectious organisms. In contrast, in the adaptive immune system, the T-cell and B-cell receptors are somatically generated in a way that gives each lymphocyte a unique structural receptor. Because the T- and B-cell receptors are not encoded in a germline, they are not predetermined to recognize any particular antigen. No matter how useful these “receptors” become, they cannot be passed down to the next generation. Although there are potentially a large number of variants of germline-encoded receptors (perhaps in the hundreds), there are 1014 and 1018 different somatically generated immunoglobulin receptors and T-cell receptors, respectively.1 Microbes are heterogenous and can mutate at very high rates. This can be handled appropriately by the adaptive immune system. This heterogeneity, however, represents more of a challenge for the innate immune system. As such, a different strategy, phylogenetically older than the adaptive system, evolved. The innate immune system has developed receptors that recognize “pathogen-associated molecular patterns” (PAMPs). PAMPs are highly conserved structures present in a large group of microorganisms. The common features of PAMPs are (1) PAMPs are only produced by microbial pathogens, not hosts; (2) structures recognized by the innate immune system are usually essential for survival or pathogenicity of the microorganism; and (3) PAMPs are usually invariant structures shared by an entire class of pathogen. The best known examples of PAMPs are bacterial lipopolysaccharide (LPS), peptidoglycan, lipoteichoic acid (LTA), mannans, bacterial DNA, double-stranded RNA, and glucans.2
Pattern Recognition Receptors
Toll-Like Receptors
At present there are nine human TLR receptors.8 The TLRs recognize molecules common to pathogens ranging from protozoa to bacteria to fungi and to viruses. The TLRs that recognize bacterial cell wall components (TLR1, -2, -4, and -6) are located on the cell surface, forming homodimers and heterodimers. Once ligated, they then translocate to the endosome for signaling (see Chapter 103).9 TLR3, TLR7, and TLR9 reside in the endosome, recognize nucleic acids produced by viruses and bacteria, and thus are available for activity against intracellular viruses.10
There are several other classes of PRRs in addition to TLRs: RIG-I–like receptors (RLRs), Nod-like receptors (NLRs), and C-type lectin receptors (CLRs) (previously described). RLRs, an intracellular receptor, recognize single- and double-stranded viral RNA and transmit their signal through a common adaptor protein, interferon promoter stimulator-1 (IPS-1) and NF-κB, to induce type I interferon production and antiviral responses.10 Double-stranded RNA-dependent protein kinase (PKR) is a unique PRR in that it can turn off protein translation directly; its effect does not seem to be modulated by downstream effectors.14 Nod1 and Nod2 are NLRs that recognize PAMPs derived from the bacterial cell wall. Nod1 and Nod2 are localized to the cytoplasm and can elicit TLR-independent antibacterial responses. Other NLRs, such as NALP1, NALP3, IPAF, and NAIP5, are components of a molecular complex called the inflammasome. The inflammasome complex comprises one or some of the NLR proteins and caspase-1. Caspase is activated in this complex and cleaves critical inflammatory molecules such as pro-IL-1β and pro-IL-18 to produce mature proteins.10 The NALP3 inflammasome is activated by stimuli such as uric acid crystal, silica, and asbestos; thus NALP3 appears to be a receptor for danger-associated molecular patterns. Infections with the malaria parasite or the fungus Candida albicans were also reported to activate the NALP3 inflammasome.
What follows is a focus on the pathogen ligands of the PRRs, in particular TLR4. It had been recognized for a number of years that CD14, present on monocytes and blood-derived macrophages, and lipopolysaccharide binding protein (LBP), found in plasma, were critical for an LPS response. However, CD14 is a glycosylphosphatidylinositol-anchored protein lacking transmembrane and intracellular domains and would be unable to signal intracellular processes. CD14 is also present to a limited extent on neutrophils.15 In addition, a soluble form of CD14 could substitute for membrane-bound form of CD14 in LPS-mediated signaling. Thus it was clear that at least one other molecule present on the cell surface would be needed to elicit an LPS-dependent response. TLR4 knockout mice and human TLR4 mutations conferred such a role for this receptor for LPS-induced responses in humans and mice. TLR4 requires an additional molecule, MD-2, that is part of the complex on the cell surface. LPB is an acute phase reactant.2,16 It catalyzes the transfer of LPS to CD14, which is then able to interact with TLR-4 and MD-2, resulting in intracellular signaling through the Toll/IL-1 receptor homologous region (TIR) adaptor molecules.9 Downstream signaling events result in release of transcriptional activating factors including NF-κB, that guide gene transcription of many genes, including TNF and interferon-β (IFN-β), a type I interferon. TNF is discussed at greater length later in this chapter; however, it is a central mediator in innate immune response pathway. IFN-β is critical in adaptive and antiviral immunity (see Chapter 103 for a description of other TLRs).
Soluble Components of Immunity
Complement System
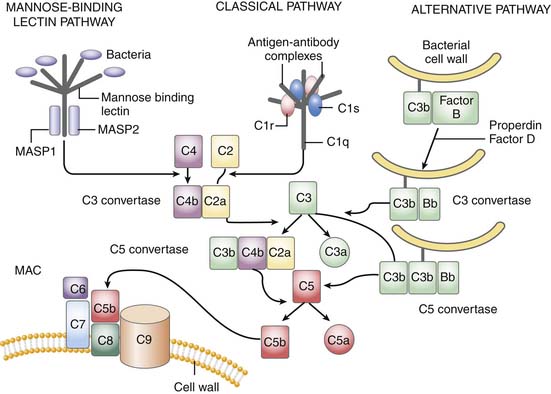
The complement system is critically positioned to participate in both the innate and adaptive immune response.26 However, it is also critical for the disposal of immune complexes from tissues and clearance of apoptotic cells.4 The three pathways in which complement may become activated are the classic, alternative, and mannose-binding lectin pathway. Although complement activation initiates differently in each, all three converge at the cleavage of C3 (Figure 91-1). The classic pathway is initiated by the binding of the C1 complex (consisting of C1q, C1r, and C1s) to antibodies bound to antigen on the cell wall. The mannose-binding lectin pathway is initiated by binding of the complex of mannose-binding lectin and the mannose-binding lectin-associate proteases 1 and 2 (MASP1 and MASP2) to arrays of mannose groups on the surface of the bacterial cell wall. Both MASP2 and C1q then function similarly in forming the C4bC2a complex, representing the convertase for C3. Bacterial products, LPS, yeast cell wall particles, and aggregated antibody, including immunoglobulin (Ig)A and IgE, are all capable of activating the alternative pathway. The alternative pathway is initiated via low-grade cleavage of C3 in plasma to C3b. C3b binds to the hydroxyl groups on cell surface carbohydrates. C3b binds factor B to form a C3bB complex, which is activated by factor D, forming C3bBb and stabilized with properdin. C3bBb then functions as an alternative convertase for C3, that cleaves many molecules of C3 to C3b. C3b binds to hydroxyl groups on the microorganism around the area of complement activation. C3a, an anaphylatoxin, is released by the C3 convertase. C3b can also bind to the C3 convertase to form C5 convertase. The C5 convertase releases the anaphylatoxin C5a and initiates the formation of the membrane attack complex (MAC), C5b6789.4 The membrane attack complex created inserts into the cell membrane, creates large pores, and leads to osmotic lysis of the target.
Thus the complement system amplifies the initial response to the organism. In addition to lysing the target organism, opsonization with complement fragments C3b/C4b and C3bi (fragment C3b) occurs, resulting in phagocytosis by neutrophils, monocytes, and macrophages. Binding occurs through specific complement receptors, CR1 for C3b/C4b (CD35) and CR3 for C3bi (CD11b/CD18, also known as Mac-1 and αMβ2). The anaphylatoxins C3a, C4a, and C5a produced are low molecular weight, biologically active peptides defined by their actions on small blood vessels, smooth muscle, mast cells, and peripheral blood leukocytes. Blood vessels, smooth muscle cells, basophils, and mast cells respond to all three anaphylatoxins. Neutrophils, monocytes, and macrophages respond to C5a. The anaphylatoxins promote edema and increase vascular permeability through the release of histamine from mast cells and through the local production of vasodilatory prostaglandins such as prostaglandin E2 (PGE2) and edemogenic leukotrienes (LT) C4, D4, and E4. C5a is a powerful activator of granulocyte function including chemotaxis, degranulation, and increased oxidative metabolism. C5a actions occur through its seven-transmembrane–spanning, G protein–coupled receptor, C5aR.27 In animal models, C5a/C5aR is critical to the development of sepsis and multiple organ failure, and potentiates many early response cytokines and coagulation. C5aR is present not only on leukocytes, but also other tissue, including the brain, kidney, gastrointestinal tract.27,28
Complement activation also occurs after oxidative stress such as occurs with ischemia/reperfusion. Complement activation is an early event after injury and the inhibition of complement activation or its components offer tissue protection after reperfusion.29 Finally, complement proteins transduce various cell signals. Complement can activate B and T cells. It can regulate apoptosis of various cell types (see Chapter 100).20,26
Immunoglobulin
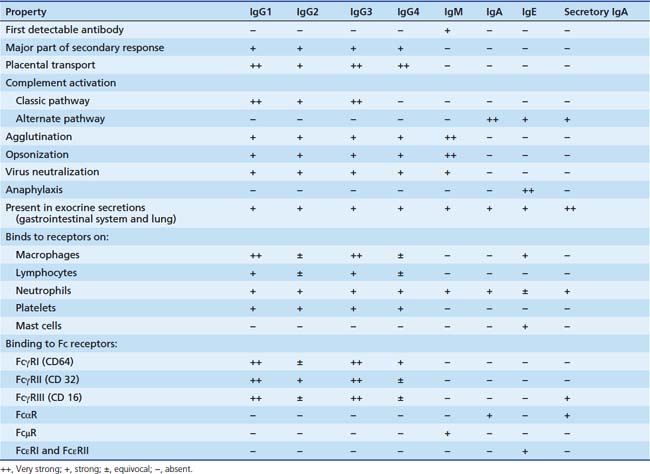
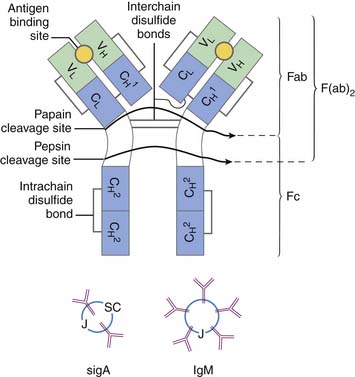
The different immunoglobulins (Igs) secreted by B cells (IgG, IgA, IgM, IgD, and IgE) are known as isotopes. IgD plays little role, if any, in containment of microorganisms. Ig isotypes can be divided into subclasses, variants that show slight structural differences but are sufficiently alike structurally to be essentially identical to other members of the isotype class. There are four IgG subclasses: IgG1, IgG2, IgG3, and IgG4. As seen in Table 91-1, isotype subclasses have specialized roles in the immune response. For example, IgG2 has a major role in the formation of carbohydrate antibodies, but has poor complement fixation characteristics. The antibody system consists of Ig present in serum, protecting the blood and tissue spaces, and that present in the secretory system, lining the gastrointestinal and respiratory tracts and present in tears. The serum component is mostly IgG (85%), with lesser amounts of IgA and IgM. The secretory system consists mostly of secretory IgA (85%), which is structurally different than serum IgA, and lesser amounts of IgG and IgM. All Igs have a basic four-chain structure composed of an identical pair of heavy (H) and light (L) chains. The H-L pairs are held together by interchain disulfide bonds and noncovalent forces. The binding site for antigen is formed by one H and one L chain. IgM is a polymer of five four-chain units, and secretory IgA is a dimer. Polymeric forms (Figure 91-2) possess a J chain that is synthesized with the H and L chains and serves to stabilize the sulfhydryl groups during polymerization. At regular intervals along the peptide chain a disulfide bond forms an intrachain loop, known as the Ig domain. This motif is repeated among immunoglobulins, T-cell receptors, adhesion proteins, and histocompatibility antigens. Proteins with these Ig domains share close homology structurally and functionally and suggest a common evolutionary origin. They are termed members of the Ig superfamily (IgSF), and all are involved in cell interaction processes associated with recognition.
The Ig molecule is divided into regions, the amino acid sequence of which is similar, such as regions needed for complement fixation or attachment to receptors on leukocytes. However, other regions are highly variable; those that bind to antigen have the highest divergency and are collectively known as the hypervariable region. Thus each Ig chain can be divided into constant and variable regions. The H chain has three constant regions and one variable region; the L chain has one variable region and one constant region. The hypervariable region of the L and H chain is tightly apposed and forms the combining site for antigen (see Figure 91-2). The digestion of Ig with papain and pepsin generates fragments with varying biologic capability. The Fc region of the Ig molecule accounts for its isotypic biologic capability. This is the region critical for complement fixation and recognition by Fc receptors on the leukocytes. The Fab region provides for specific unique antigen-antibody interactions. The F(ab′)2 fragment is formed with pepsin cleavage; the affinity for antigen is twice as great as Fab alone.
Contact Activation System
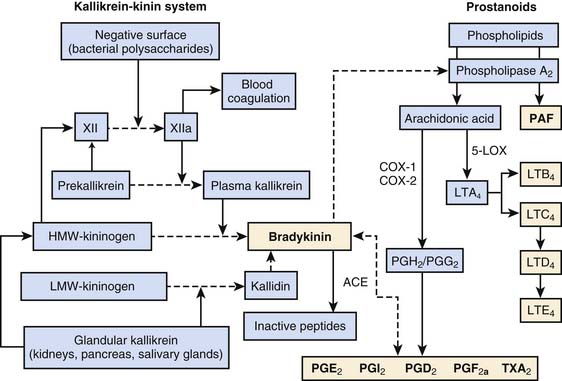
There are four major plasma protein systems that contribute to the host’s defense and participate in the development of inflammatory tissue injury: the complement system (previously discussed), the contact activation system (also known as Hageman factor or intrinsic coagulation system), the extrinsic coagulation cascade, and the fibrinolytic system (see Chapter 80). The contact activation system is critical to host defense and control of local blood flow at sites of injury. Hageman factor (Factor XII) is activated spontaneously (XIIa) on contact with negatively charged surfaces, such as lipid A of LPS and vascular basement membranes (Figure 91-3). High-molecular-weight kininogen (HMWK), prekallikrein, and factor XI circulate in the plasma as complexes. Factor XIIa will activate factor XI and cleave prekallikrein to kallikrein. Kallikrein will then cleave HMWK to bradykinin. The kallikrein-kinin system also encompasses the tissue (or glandular) kallikrein-kinin system (see Figure 91-3). Tissue kallikrein is immunologically distinct from plasma kallikrein and present throughout the body as an inactive “pro-” substance. In the presence of intracellular enzymes, plasmin, or plasma kallikrein, tissue kallikrein is produced, secreted, and active in the tissue where it is made. Tissue kallikrein can then cleave HMWK to bradykinin directly, or it can cleave low-molecular-weight kininogen (LMWK) or tissue kininogen (T-kininogen) to kallidin that is then directly converted to bradykinin. In the plasma, 80% of kininogen is low molecular weight.
Bradykinin is an exceedingly potent vasoactive peptide.31 It can cause venous dilation, increased vascular permeability, hypotension, bronchoconstriction, and activation of phospholipase A2. Phospholipase A2 releases arachidonic acid from cell membrane and initiates the production of both proinflammatory and antiinflammatory phospholipids-derived products (see subsequent section and Figure 91-3). Bradykinin is metabolized by angiotensin-converting enzyme (ACE) to inactive peptides. Bradykinin, along with prostanoids, stimulates the pain response through polymodal receptors and C-fibers (capsaicin sensitive).32 Bradykinin has been shown to be responsible for the four signs of inflammation: heat, redness, swelling, and pain. The bradykinin effect is enhanced by simultaneous production of prostanoids. Prostanoids, particularly along with the low pH of exudates, inhibit the activity of kininases such as ACE.
Lipid-Derived Mediators of Inflammation
Although these products per se are not “soluble” components of immunity, their production nonetheless has both proinflammatory and antiinflammatory effects. They are discussed here because of their production as a result of the contact activation syndrome. These mediators are not stored preformed, but are rapidly generated after cell stimulation. Arachidonic acid (AA, or eicosatetranoic acid) is a 20-carbon fatty acid. Its metabolites are termed eicosanoids. The eicosanoids are produced by a variety of cell type–, tissue-, and species-specific biosynthetic pathways. Prostanoids are a specific class of mediators generated via initial actions of cyclooxygenase. The eicosanoid family includes the thromboxanes, prostacyclins, leukotrienes, hydroeicosotetranoic acids (HETES), epoxyeicosatrienoic acids (EETs), lipoxins, and isoprostanes. Release of AA and the 1-alkyl-2-acetyl analogs of phosphatidyl choline (platelet activating factor [PAF]) occurs through the action of phospholipase A2 and phospholipase C on cell membrane phospholipids. Phospholipases can be stored in lysozymes or exist in the cytoplasm. Through cell activation, they translocate to the inner cell membrane, where they can hydrolyze cell membrane phospholipids. The major routes of AA metabolism for proinflammatory effects are the 5-lipoxygenase pathway (production of the leukotrienes, LT) and the cyclooxygenase pathway (production of prostaglandins and thromboxane, i.e., prostanoids). Of particular interest are newly recognized classes of prostanoids derived from polyunsaturated fatty acids (omega-3 fatty acids) that mediate antiinflammatory effects, the resolvins and protectin.14
Cytokines
Cytokines are signaling proteins secreted by cells that affect the functional properties of other cells of the same organism. The cytokine family includes lymphokines, chemokines, interleukins, and interferons. Unlike circulating hormones, cytokines travel short extracellular distances before interacting with target cell surface receptors in a paracrine or autocrine manner. Cytokines can be detected in serum samples, particularly during times of maximal production, as occurs in sepsis. Cytokines as a group are low-molecular-weight (<80 kDa) proteins. They interact with high-affinity cell surface receptors specific for each cytokine. Their cell surface binding ultimately leads to changes in the pattern of protein synthesis and/or altered cell behavior. They often have multiple-overlapping cell regulatory functions. Many cytokines are produced early in infection, whereas others are produced at later stages.
Interleukin-1 and Tumor Necrosis Factor
IL-1 is a phylogenetically old molecule that predates the evolution of lymphocytes and immunoglobulin. Its activity extends beyond immune function. IL-1 is produced by a wide variety of cells, including macrophages, endothelial cells, epithelial cells, and vascular smooth muscle cells. There are two separate forms of IL-1, IL-1α, and IL-1β. In contrast, TNF-α is produced by cells primarily of the innate immune system, including monocytes/macrophages, NK cells, mast cells, and neutrophils under specific conditions. TNF is also produced by other cell types under conditions of stress. For example, TNF is produced by cardiac myocytes and is implicated in both acute and chronic congestive heart failure as well as in the cardiomyopathy associated with sepsis. TNF-β (also known as lymphokine) is produced by T lymphocytes, but occasionally antigen-activated T cells may also produce TNF-α. Both TNF and IL-1 are produced as small precursor molecules or “pro” molecules that are cleaved by IL-1β converting enzyme in the inflammasome (by caspase 1) and TNF-α converting enzyme, ADAM17 (a disintegrin and metalloproteinase), respectively. Once cleaved, these proteins are then excreted. It should be noted that caspase-1 (in the inflammasome) and ADAM17 are multifunctional proteinases and have activity on other interleukins and inflammatory molecules. IL-1 is the only cytokine with a natural inhibitor, IL-1 receptor antagonist (IL-1RA) produced by the same cells that produce IL-1. IL-1RA functions to downregulate the proinflammatory effects of IL-1. IL-1 and TNF-α are the early major mediators of gram-negative endotoxin shock (see Chapter 103). As outlined earlier in this chapter, signaling of cells by TNF and IL-1 occurs at least in part through NF-κB, and consequently share a similarity in receptor function and signaling molecules.
IL-2 is produced by CD4 T cells when they encounter foreign peptide-MHC complex on antigen-presenting cells. This cytokine is critical for adaptive immune response in that it has both an autocrine and paracrine function, triggering T cells to undergo multiple rounds of proliferation and differentiate into effector T cells.
Interferons and Other Soluble Products
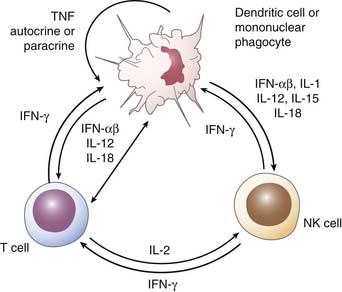
IFN-α and IFN-β are known as type I interferons. IFN-α is produced by monocytes and macrophages, whereas IFN-β is produced by fibroblasts and other cell types. The major stimuli for type I interferon are viral infections; they also respond to T-cell–derived factors in adaptive immune responses. Type I IFNs inhibit viral replication, and patients with insufficient production suffer from severe progressive or fulminant viral disease. These IFNs inhibit cell proliferation, enhance the lytic potential of NK cells, and increase class I HLA while decreasing class II HLA. IFN-γ is a type II IFN and is produced primarily by CD4+, CD8+, and NK cells (Figure 91-4). IFN-γ has antiviral and antiproliferative activity. It upregulates class I and II HLA expression, thus enhancing cellular toxicity and antigen presentation, respectively. IFN-γ activates monocytes and macrophages as well as neutrophils, resulting in enhanced killing of intracellular organisms, including mycobacteria and listeria. In addition, IFN-γ induces inducible nitric oxide synthase in macrophages, resulting in the generation of nitric oxide, a critical component for bactericidal function. Animals with a deficiency of IFN-γ have decreased survival in response to salmonella and mycobacterial infections. Patients with complete loss of IFNγ receptors have severe, early life infections with salmonella and viral infections (including respiratory syncytial virus, parainfluenza, herpes simplex virus, and cytomegalovirus) with a high mortality rate.35,40,42 IFN-γ is a critical cytokine at the interface of the adaptive and innate immune system because of its function on NK cells and ultimately monocytes/macrophages.
Chemokines are structurally and functionally related inflammatory cytokines with the ability to stimulate the chemotactic migration of distinct sets of cells, including neutrophils, monocytes, lymphocytes, dendritic cells, macrophages, fibroblasts, stem cells, and smooth muscle cells. The chemokine family is the largest family of cytokines, and although their main function is characterized as “chemotaxis,” or directing migration through a concentration gradient, they also have a number of other functions, including cell activation, signaling, effects on angiogenesis and tumorigenesis, as well as immune cell polarization. To date there are 40+ identified chemokines.49 They are small (∼8 to 14 kDa), mostly basic molecules. They function through unique receptors that are G protein coupled and are seven membrane spanning (i.e., have seven transmembrane domains).50 The cytoplasmic domains of the receptors are critical for cell signaling and function. Chemokines may use more than one receptor for function. Chemokines are central to the process of extravasation of leukocytes that includes multiple steps involving interactions of adhesion molecules and the chemoattractant function of these proteins.51 Chemokines are defined by structure, not function, and can be divided into two large and two small subgroups depending on the number and arrangement of conserved cysteines. The subgroups are CC, CXC, C, and CX3C. It should be appreciated that in the past 10 years, chemokines have been designated by their subgroup followed by ligand number; for example, IL-8 is CXCL8. Most chemokines are classified into two main groups according to function: (1) those concerned with hemostasis that are constitutively expressed and coordinate leukocyte trafficking during hematopoiesis and those with lymphocyte recirculation, and (2) those concerned with inflammation and tissue injury. Chemokines are produced by hematopoietic cells themselves as well as by endothelial cells, epithelial cells, and cells arising from the mesoderm, including fibroblasts, myocytes, hepatocytes, and lymphatic cells. Table 91-2 lists a select group of cytokines involved in infection and inflammation.
Table 91–2 Chemokine/Chemokine Receptor Families (Partial List)
| Name | Original Ligand Name∗ | Chemokine Receptor |
|---|---|---|
| CXCL1 | GRO-α/MGSA-α | CXCR2>CXCR1 |
| CXCL2 | GRO-β/MGSA-β | CXCR2 |
| CXCL3 | GRO-γβ/MGSA-γ | CXCR2 |
| CXCL5 | ENA-78 | CXCR2 |
| CXCL7 | NAP-2 | CXCR2 |
| CXCL8 | IL-8 | CXCR1, CXCR2 |
| CXCL9 | Mig | CXCR3 |
| CXCL10 | IP-10 | CXCR3 |
| CXCL12 | SDF-1 | CXCR4 |
| C CHEMOKINE/RECEPTOR FAMILY | ||
| XCL-1 | Lymphotactin | XCR1 |
| CX3C CHEMOKINE/RECEPTOR FAMILY | ||
| CX3CL1 | Fractalkine | CX3CR1 |
| CC CHEMOKINE/RECEPTOR FAMILY | ||
| CCL2 | MCP-1 | CCR2 |
| CCL3 | MIP-1α | CCR1,CCR5 |
| CCL4 | MIP-1β | CCR5 |
| CCL5 | RANTES | CCR1, CCR3, CCR5 |
| CCL8 | MCP-2 | CCR3 |
| CCL11 | Eotaxin | CCR3 |
| CCL19 | MIP-3β | CCR7 |
| CCL21 | 6Ckine | CCR7 |
| CHEMOKINE RECEPTORS AND CELLULAR DISTRIBUTION | ||
| XCR1 | Lymphotactin | T,B, NK |
| CXCR1 | IL-8, GRO-α | N,M, T, NK, En, Ms, Bs |
| CXCR2 | IL-8, GRO-α,-β-γ, NAP-2, ENA-78 | N,M,T,NK, Ms, As, Nn, En |
| CXCR3 | IP-10, Mig | Activated T |
| CXCR4 | SDF-1 | Myeloid, T, B, Ep, En, DC |
| CX3CR1 | Fractalkine | NK, M, T |
| CCR1 | RANTES, MIP-1α, MCP-2, MCP-3 | N, M, T, NK, B, Ms, As, Nn |
| CCR2 | MCP-1 | M, T, B, Bs |
| CCR3 | RANTES, eotaxin | Eo, Bs, T |
| CCR5 | RANTES, MIP-1α, MIP-1β, MCP-2 | T, M, Mφ |
| CCR7 | MIP-3β, 6Ckine | T, B, DC |
GRO, Growth regulating peptide; MGSA, melanocyte growth stimulating activity; ENA, epithelial-derived neutrophil attractant; NAP, neutrophil activating peptide; IL-8, interleukin-8; IP-10, γ-interferon-induced peptide 10; SDF, stroma derived factor; MCP, monocytes chemotactic peptide; MIP, macrophage inflammatory peptide; RANTES, regulated on activation, normal T cell expressed and secreted; T, T cell ; B, B cell; NK, natural killer cell; M, monocyte/macrophage; N, neutrophil; Ms, mast cell; Bs, basophil; As, astrocyte; Nn, neuron; En, endothelium, Eo, eosinophils; DC, dendritic cell; Ep, epithelial cell; Mφ macrophage.
∗ All ligand names are human chemokines.
Modified from Murdoch C, Finn A: Chemokine receptors and their role in inflammation and infectious diseases, Blood 95:3032, 2000; and Zlotnik A, Yoshie O: Chemokines: a new classification system and their role in immunity, Immunity 12:121, 2000.
Cellular Components of Immunity
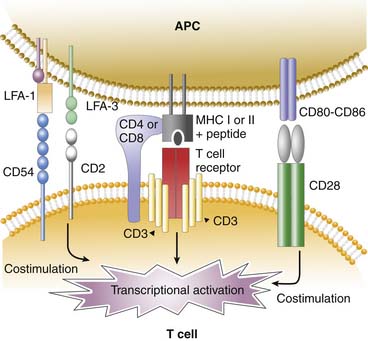
The major T-cell subsets are the CD4+ helper/inducer and the CD8+ suppressor/killer cells. Nearly all T cells bear a T-cell receptor composed of an α- and β-chain; they also express CD4 or CD8 coreceptors. Nearly all the αβ T cells recognize protein antigen in the form of peptide fragments bound to classic MHC molecules (MHC class I or MHC class II). The CD4 molecule augments binding to antigens presented in association with MHC II antigens, whereas CD 8 molecules are necessary for antigen binding to MHC I. The immunologic synapse is the highly ordered junction that forms between the APC, such as the dendritic cells or tissue macrophages, and the T cell during antigenic stimulation. The structure resembles a doughnut in which the T-cell receptor-peptide-MHC, CD3, CD4, or CD8 is in the center of the synapse. The costimulatory molecules CD2, CD28, CD54 (intercellular adhesion molecule-1 [ICAM-1]) on the T cell bind to their respective ligands LFA-3 (CD 58), CD80-CD86, LFA-1 (CD11a/CD18) on the APC on the perimeter of synapse (Figure 91-5).59
The CD4+ naive cells, which are those never exposed to antigen, can differentiate into effector cells expressing specific patterns of cytokines. Originally designated as Th1 and Th2 based on the cytokines they produce, there has been an explosion in the past 15 years delineating additional subtypes. Most of these cytokines produced by T cells are secreted, but some can be expressed on the cell surface. IL-12 and IFN-γ drive T cells into the Th1 pathway. IFN-γ is the signature cytokine produced by Th1 cells, but the Th1 cells also produce substantial amounts of IL-2, TNF-α, and TNF-β. The Th1 response is considered proinflammatory. IL-4 drives T cells into the Th2 pathway. Th2 cells also produce large amounts of IL-4 along with IL-5, IL-9, and IL-13. IL-4 and IL-13 (along with IL-10 produced by monocytes/macrophages) are considered antiinflammatory or immunosuppressive. The Th2 response is critical for eosinophil function and is important in the development of IgE responses and the killing of parasites.
TGF-β is produced by many cells. In the absence of IL-6, it will induce CD4+ cells into a regulatory T cell, known as a T-reg. T-reg cells are identified by a transcription factor, Foxp3+ (Forkhead box P3 transcription factor) and carry the IL-2 receptor CD25. Presence of Foxp3+ is implicated in the development of autoimmunity, allergy, and rejection in transplant medicine and suppression of immune responses to cancer.60 There are several different types of T-regs. All require cell-to-cell contact for immune suppression.
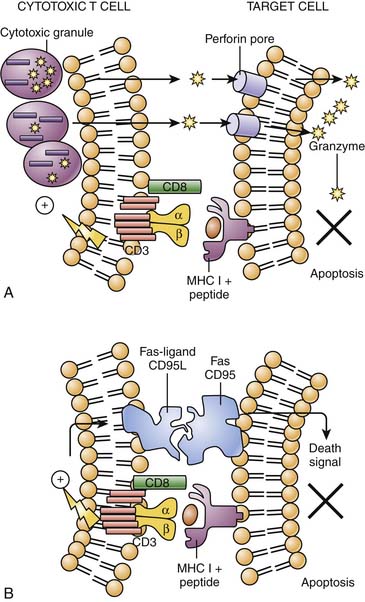
Naive CD8+ cells are not effective killer cells. However, after activation with antigen in the context of MHC class I by APCs in the presence of IL-2 and IL-12, they differentiate quickly into CD8+ cytotoxic cells. These cells express perforin, granzymes, and Fas ligand and produce effector cytokines including TNF-α and IFN-γ. Perforins introduce pores into the target cell through which granzymes can enter, leading to the triggering of apoptosis and cell death. Alternatively, the cytotoxic T cell upregulates the Fas ligand (CD95L) that engages Fas (CD95) on the target cell, resulting in delivery of death signal culminating in apoptosis (Figure 91-6).
NK cells are large granular lymphocytes with innate immune function. They play a critical role in the early host defense against viral, bacterial, and other infections as well as cancer. NK cells recognize their targets through unique NK receptors and are able to recognize self-MHC class I or class I–like molecules that inhibit or enhance NK function. Their phenotype is characterized by the expression of the CD56 surface antigen and the lack of CD3. NK cells produce IFN-γ, TNF-α, IL-10, and GM-CSF. They exhibit spontaneous cytotoxic activity against virus-infected cells and mediate antibody-dependent cell cytotoxicity through FCRγIII (CD16). Cytotoxicity is the major effector function of NK cells. They bridge the innate and adaptive immune response (Figure 91-6).62,63 The NKT cells express both CD56 and CD3 T-cell receptor and thus share receptor structures of both conventional NK and T cells. NKT cells are potentially capable of very rapid secretion of large amounts of Th1 or Th2 cytokines but also contain perforin. Both NK and NKT cells use the same mechanisms as the cytotoxic CD8+ T cell for killing (i.e., perforin/granzyme cytotoxicity and Fas-ligand/fas cytotoxicity) (see Figure 91-6). The control and resolution of viral infections require the eliminate of the source of the virus, hence the destruction of the virus-infected cell before progeny virus is produced. This is mediated during the innate phase of the immune response by NK cells. In most cases, resolution of active viral infection ultimately requires the development of antigen-specific T lymphocytes, the majority of which are MHC class I–restricted CD8+ T cells, although MHC class II CD4+ cells and γδ T cells may also mediate cytotoxicity.
Phagocytic cells include neutrophils, eosinophils, monocytes/macrophages, and dendritic cells. They have in common a number of different properties that are of prime importance to the host inflammatory response. Neutrophils, eosinophils, and monocytes/macrophages share the ability to phagocytose foreign material, release granule constituents, secrete inflammatory mediators and regulators, and synthesize reactive oxygen products through a unique reduced nicotinamide adenine dinucleotide phosphate (NADPH) oxidase enzyme system present on the cell membrane. Neutrophils, eosinophils and basophils are all polymorphonuclear leukocytes (PMNLs). Neutrophils and eosinophils share similar mechanisms of cell migration, phagocytosis, and pathogen killing. All three cell types have segmented nuclei and contain granules, although their granule content varies. Neutrophils are the host’s main defense against bacterial and fungal infections. Eosinophils are important to the control of parasitic infections. Neutrophils may remain in the storage pool of the bone marrow for up to 5 days. Released from the bone marrow into the blood, about half the neutrophils circulate for about 10 hours; the other half remains in a marginated pool, so they are not accessible to phlebotomy. This marginated pool is thought to be in the spleen, along vessel walls, and in the lung microcirculation. Cells can be mobilized from this marginated pool by infection/inflammation and stress. Once circulating neutrophils migrate into the tissue they survive for 1 to 2 days, likely longer in the presence of G-CSF and GM-CSF.
Neutrophils from patients with chronic granulomatous disease retain some of the antimicrobial activity of normal neutrophils despite the inability to produce oxygen species. This is due to the many endogenous antimicrobials present in the granules that are critical to the killing of microbes. These antimicrobials, discussed earlier in the chapter, include defensins, bacterial permeability increasing protein, lactoferrin, and lysozyme. A number of other proteases, hydrolases, and nucleases in the granules of phagocytes, although not directly microbicidal, act synergistically with the antimicrobial agents to contribute to killing. These products can also result in host injury if released from the neutrophil. Neutrophil elastase, collagenase, and gelatinase (also known as matrix metalloproteinase-8, -9, respectively) can hydrolyze key components of the extracellular matrix. Neutrophil elastase not only can degrade almost all components of the extracellular matrix, but can also cleave a variety of key plasma proteins such as immunoglobulins, complement proteins, and clotting factors. The activity of the elastase outside the cells is regulated primarily by α1-proteinase inhibitor. Neutrophil elastase can mediate injury outside the neutrophil when α1-proteinase inhibitor is inactivated by oxidants.65
Immature DCs migrate to and remain in the periphery, where they express low levels of MCH I and II. These immature DCs will then mature either after phagocytosis of foreign antigen or by activation of one of its receptors (e.g., TLR). The mature DC will then migrate to the local lymphoid organ; as they do, they increase cell surface expression of MHC as well as T costimulatory molecules (i.e., CD80-CD86) and begin to secrete specific cytokines. Mature DCs lose the ability to phagocytose. In the lymphoid organ, the DC will present antigen to the CD4+ or CD8+ T-cell receptor via MHC II or I, respectively. The DC costimulatory molecules CD80 or CD86 must engage their ligand, CD28, on the T cell for full activation (Figure 91-6). It should be noted that for the initial immune response, the DC (or APC) must be in geographic proximity of the CD8+ and CD4+ cell. Alternatively, the DC or APC may be preconditioned by an activated CD4+ cell that is then able to activate naive CD8+ cells to become cytotoxic T cells.66,67 Not all macrophages function well as antigen presenting cells; elicited peritoneal macrophages do, but alveolar macrophages and Kupffer cells do not.15
Leukocyte Localization
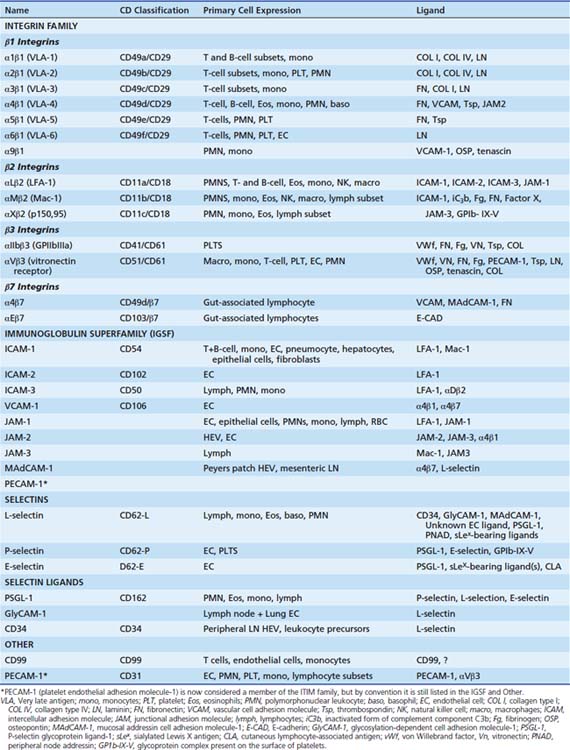
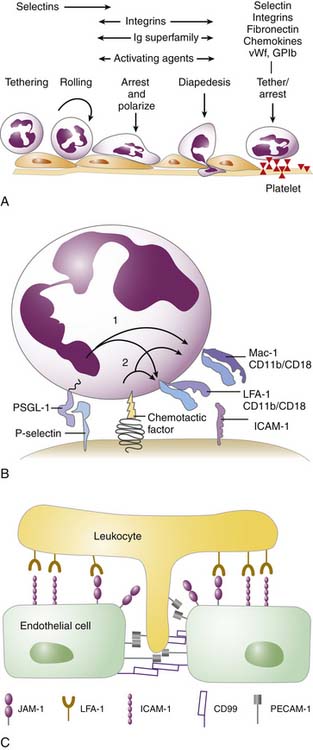
In general, the multistep process begins by activation of the postcapillary venular endothelial surface by inflammatory cytokines such as IL-1, TNF, IL-4, and IFNγ. The surface transforms from a nonadhesive surface to one that is proadhesive through the expression of specific ligands. These ligands will recognize their cognate receptors on the circulating effector leukocytes (and platelets) (i.e., neutrophils, eosinophils, monocytes, NK cells, activated T cells). Endothelial ligands that are upregulated include members of the selectin family and IgSF (Table 91-3). Selectins are responsible for the initial capture of the leukocyte from the free-flowing stream as well as rolling on the endothelial surface of the IgSF that is critical for leukocyte slowing, arrest, and migration on the cell surface (Figure 91-7). The leukocyte ligand for E- and P-selectin is PSGL-1 (CD162, P-selectin glycoprotein ligand 1). The leukocyte receptors for the IgSF are the β2 integrins, in particular Mac-1 (CD11b/CD18), LFA-1 (CD11a/CD18), and the β1 integrin VLA-4 (CD49d/CD29). The leukocyte integrins are heterodimers composed of an α and β subunit. The β subunit may be shared by multiple members of a subfamily, whereas the α subunit confers specificity. Mac-1 functions not only in leukocyte recruitment, but also as a receptor for complement fragment C3bi; hence its alternative name of complement receptor 3. LFA-1 also functions as coinducer of the immune response in the immunologic synapse as discussed above (Table 91-3). The endothelial surface also secretes a number of chemokines and other activating substances, such as PAF, that activate the leukocyte and mediate the transition from rolling to arrest. After the leukocyte has arrested, it polarizes, then “crawls” and emigrates through the endothelial lining of the vessel. This emigration, known as diapedesis, is in response to activating agents (chemokines, LTB4) released by cells present in the subendothelial matrix, released bacterial products (N-formyl peptides), or through complement activation (C5a). This process is also dependent on leukocyte integrins that recognize IgSF on the endothelial cells necessary for transendothelial migration (see Figure 91-7). Locomotion through the subendothelial matrix requires additional leukocyte integrins (VLA-1, -2, -3, 5, -6) that recognize matrix proteins, including fibronectin, collagen, vitronectin, and vimentin. What effector cell is recruited and the tissue to which it is recruited depend on the adhesion molecules present on the endothelial surface and the effector cells as well as the “signals” released or presented at the endothelial surface and in the subendothelial region.71-73
This multistep paradigm is also critical for the homing of lymphocytes to the high endothelial venules, a specialized endothelium of the secondary lymphoid organs. Here naive lymphocytes (rather than activated cells) tether, roll, arrest, and emigrate through the endothelial surface. The naive lymphocytes migrate toward DC or APC that secrete cytokines and chemokines necessary for activation of the T cell through the T-cell receptor.74 One critical chemokine is SDF-1 (CXCL12). Its receptor, CXCR4, is present on almost all lymphocytes and many monocytes. The multistep paradigm is not operative in all vascular beds. In lungs the inflammatory cells do not “roll,” but instead are physically trapped in the pulmonary capillary bed and emigrate from this area rather than in the postcapillary venules.75 In the liver, leukocytes are physically trapped in the sinusoids. Because the sinusoidal endothelia has large pores, the leukocytes can easily interact with the underlying hepatocytes76 The platelet can also function as a surface to which a leukocyte can bind by the activated release from the Weibel-Palade body of P-selectin at sites of injury or inflammation to the endothelium (see Figure 91-7).
Host Response to Infection: A Summary
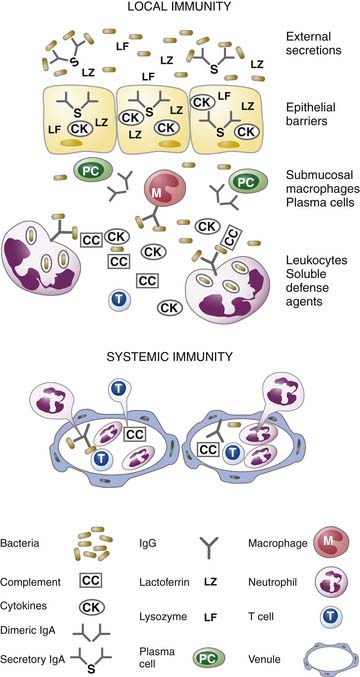
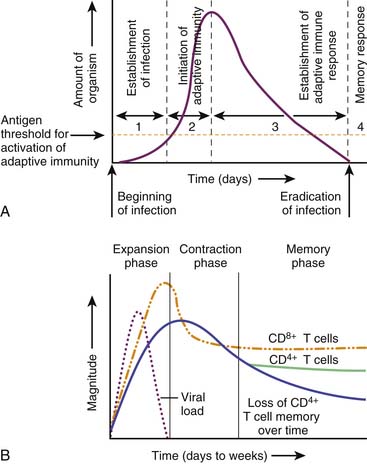
There is a coordinated and highly regulated response by the body to microbial infection. As outlined in Figure 91-8, the first defense is local immunity. The epithelial surface functions as a physical barrier. Through the release of antimicrobial peptides from the epithelium and secretory IgA from submucosal plasma cells, microbial burden is decreased. The epithelial cells at the site of infection will produce cytokines and chemokines that regulate the invasion of the area by leukocytes. However, these cytokines will also modulate the submucosal macrophages and plasma cells that are constitutively present. The presence of soluble agents critical for opsonization (including Ig and complement components) permit the efficient phagocytosis of organisms by recruited neutrophils and monocytes as well as resident macrophages. The presence of T cells and plasma cells early in the infection depends on a previous encounter with the organism. If the host has immunologic memory for the microbe, then there will be fairly rapid (i.e., within 1 day) expansion of the memory T cells to effector cells (i.e., cytotoxic T cells) and expansion of memory B cells and differentiation to plasma cells to produce IgG. It should be noted that at each step of the process, the number of bacteria decrease.76,79
If the host must mount a primary immune response to control and eliminate the infection, it may take 3 to 5 days for microbial or viral elimination to occur if local mechanisms or innate immune systems response are inadequate. As outlined in Figure 91-9, the initiation of adaptive immunity for a typical viral infection occurs within 2 days. Between 3 and 4 days, there is the establishment of the adaptive immune response with clonal expansion of CD8+ and CD4+ effector cells, and eradication of the infection occurs. After eradication of the infection there is contraction of the clonal response, but the continued presence of antibody, residual effector cells, and immunologic memory provide lasting protection against reinfection.
Apoptosis is the process of programmed cell death necessary for the resolution of the inflammatory response (see Chapter 100). As cells begin to die, they express ligands on their surfaces that signal resident tissue macrophages for removal via phagocytosis. During apoptosis the cell’s nucleus and cytoplasm condense, nuclear DNA is degraded into small dense pieces, and marked cytoplasmic vesiculation and blebbing of the plasma membrane occur. In the final stages the cell collapses into multiple fragments (apoptotic bodies). Apoptosis can be triggered by external forces such as interaction of CD95 ligand (Fas ligand) on cytotoxic T lymphocytes or NK cells with CFD95 on a target cell such as a virally infected cell (see Figure 91-6). Apoptosis can also be initiated by signals arising from DNA or mitochondrial damage.80,81 The signals converge on activation of a family of proteases called caspases that cleave multiple substrates to induce destruction of the cell from within. This process is central to peripheral deletion of excess T and B lymphocytes as the immune response wanes, to clearances of infected cells, and to resolution of inflammation with the removal of emigrated leukocytes.82 The final step of apoptosis is the removal via the tissue macrophage.83 In the process of eliminating apoptotic cells, the macrophage produces “immunosuppressive” cytokines such as TGF-β and other cytokines that downregulate the inflammatory response (e.g., IL-10).
References are available online at http://www.expertconsult.com.