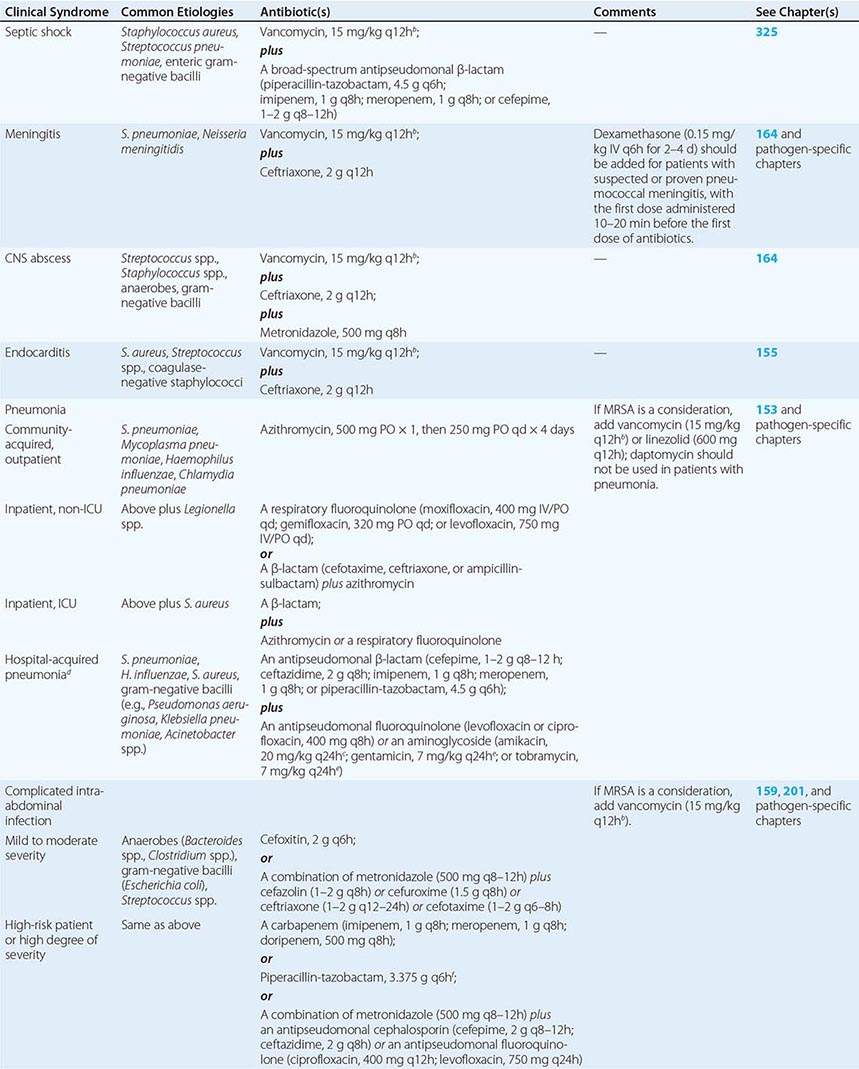
PART 8: Infectious Diseases
SECTION 1 |
BASIC CONSIDERATIONS IN INFECTIOUS DISEASES |
144 |
Approach to the Patient with an Infectious Disease |
HISTORICAL PERSPECTIVE
The origins of the field of infectious diseases are humble. The notion that communicable diseases were due to a miasma (“bad air”) can be traced back to at least the mid-sixteenth century. Not until the work of Louis Pasteur and Robert Koch in the late nineteenth century was there credible evidence supporting the germ theory of disease—i.e., that microorganisms are the direct cause of infections. In contrast to this relatively slow start, the twentieth century saw remarkable advances in the field of infectious diseases, and the etiologic agents of numerous infectious diseases were soon identified. Furthermore, the discovery of antibiotics and the advent of vaccines against some of the most deadly and debilitating infections greatly altered the landscape of human health. Indeed, the twentieth century saw the elimination of smallpox, one of the great scourges in the history of humanity. These remarkable successes prompted noted scholar Aidan Cockburn to write in a 1963 publication entitled The Evolution and Eradication of Infectious Diseases: “It seems reasonable to anticipate that within some measurable time … all the major infections will have disappeared.” Professor Cockburn was not alone in this view. Robert Petersdorf, a renowned infectious disease expert and former editor of this textbook, wrote in 1978 that “even with my great personal loyalties to infectious diseases, I cannot conceive a need for 309 more [graduating trainees in infectious diseases] unless they spend their time culturing each other.” Given the enormous growth of interest in the microbiome in the past 5 years, Dr. Petersdorf’s statement might have been ironically clairvoyant, although he could have had no idea what was in store for humanity, with an onslaught of new, emerging, and re-emerging infectious diseases.
Clearly, even with all the advances of the twentieth century, infectious diseases continue to represent a formidable challenge for patients and physicians alike. Furthermore, during the latter half of the century, several chronic diseases were demonstrated to be directly or indirectly caused by infectious microbes; perhaps the most notable examples are the associations of Helicobacter pylori with peptic ulcer disease and gastric carcinoma, human papillomavirus with cervical cancer, and hepatitis B and C viruses with liver cancer. In fact, ~16% of all malignancies are now known to be associated with an infectious cause. In addition, numerous emerging and re-emerging infectious diseases continue to have a dire impact on global health: HIV/AIDS, pandemic influenza, and severe acute respiratory syndrome (SARS) are but a few examples. The fear of weaponizing pathogens for bioterrorism is ever present and poses a potentially enormous threat to public health. Moreover, escalating antimicrobial resistance in clinically relevant microbes (e.g., Mycobacterium tuberculosis, Staphylococcus aureus, Streptococcus pneumoniae, Plasmodium species, and HIV) signifies that the administration of antimicrobial agents—once thought to be a panacea—requires appropriate stewardship. For all these reasons, infectious diseases continue to exert grim effects on individual patients as well as on international public health. Even with all the successes of the past century, physicians must be as thoughtful about infectious diseases now as they were at the beginning of the twentieth century.
GLOBAL CONSIDERATIONS
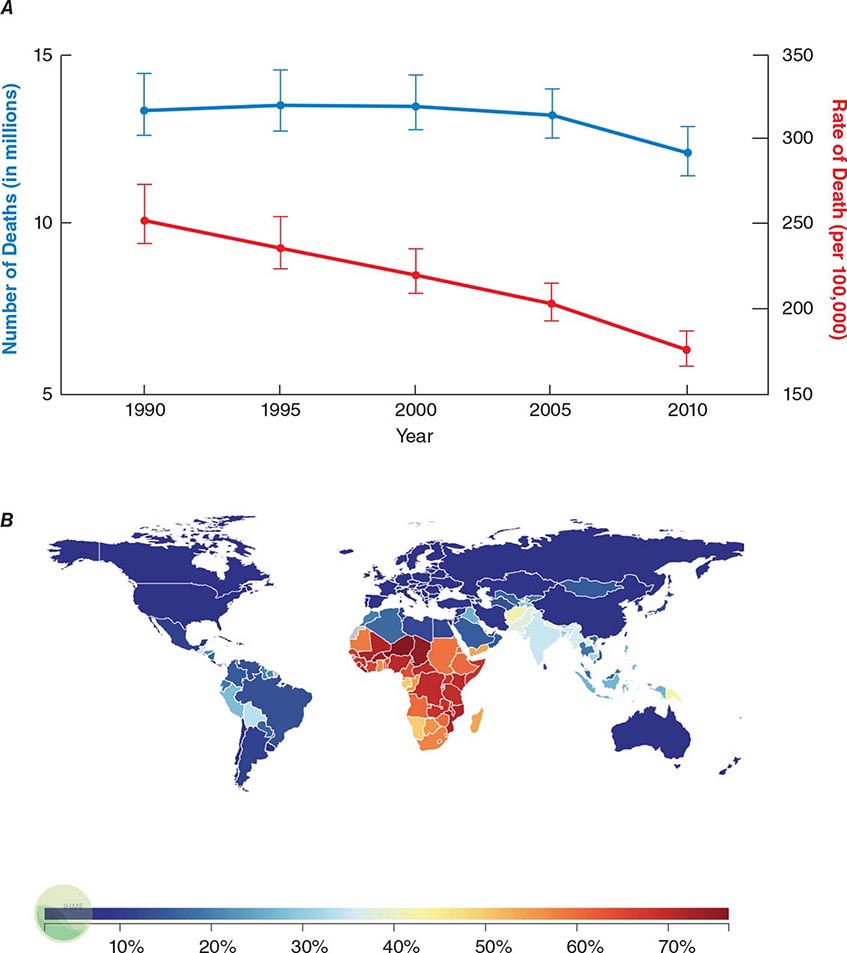
![]() Infectious diseases remain the second leading cause of death worldwide. Although the rate of infectious disease–related deaths has decreased dramatically over the past 20 years, the absolute numbers of such deaths have remained relatively constant, totaling just over 12 million in 2010 (Fig. 144-1A). As shown in Fig. 144-1B, these deaths disproportionately affect low- and middle-income countries (Chap. 13e); in 2010, 23% of all deaths worldwide were related to infectious diseases, with rates >60% in most sub-Saharan African countries.
Infectious diseases remain the second leading cause of death worldwide. Although the rate of infectious disease–related deaths has decreased dramatically over the past 20 years, the absolute numbers of such deaths have remained relatively constant, totaling just over 12 million in 2010 (Fig. 144-1A). As shown in Fig. 144-1B, these deaths disproportionately affect low- and middle-income countries (Chap. 13e); in 2010, 23% of all deaths worldwide were related to infectious diseases, with rates >60% in most sub-Saharan African countries.
FIGURE 144-1 Magnitude of infectious disease–related deaths globally. A. The absolute number (blue line; left axis) and rate (red line; right axis) of infectious disease–related deaths throughout the world since 1990. B. A map depicting country-specific data for the percentages of total deaths that were attributable to communicable, maternal, neonatal, and nutritional disorders in 2010. (Source: Global Burden of Disease Study, Institute for Health Metrics and Evaluation.)
Given that infectious diseases are still a major cause of global mortality, understanding the local epidemiology of disease is critically important in evaluating patients. Diseases such as HIV/AIDS have decimated sub-Saharan Africa, with HIV-infected adults representing 15–26% of the total population in countries like Zimbabwe, Botswana, and Swaziland. Moreover, drug-resistant tuberculosis is rampant throughout the former Soviet-bloc countries, India, China, and South Africa. The ready availability of this type of information allows physicians to develop appropriate differential diagnoses and treatment plans for individual patients. Programs such as the Global Burden of Disease seek to quantify human losses (e.g., deaths, disability-adjusted life years) due to diseases by age, sex, and country over time; these data not only help inform local, national, and international health policy but can also help guide local medical decision-making. Even though some diseases (e.g., pandemic influenza, SARS) are seemingly geographically restricted, the increasing ease of rapid worldwide travel has raised concern about their swift spread around the globe. The world’s increasing interconnectedness has profound implications not only for the global economy but also for medicine and the spread of infectious diseases.
UNDERSTANDING THE MICROBIOTA
Normal, healthy humans are colonized with over 100 trillion bacteria as well as countless viruses, fungi, and archaea; taken together, these microorganisms outnumber human cells by 10–100 times (Chap. 86e). The major reservoir of these microbes is the gastrointestinal tract, but very substantial numbers of microbes live in the female genital tract, the oral cavity, and the nasopharynx. There is increasing interest in the skin and even the lungs as sites where microbial colonization might be highly relevant to the biology and disease susceptibility of the host. These commensal organisms provide the host with myriad benefits, from aiding in metabolism to shaping the immune system. With regard to infectious diseases, the vast majority of infections are caused by organisms that are part of the normal flora (e.g., S. aureus, S. pneumoniae, Pseudomonas aeruginosa), with relatively few infections due to organisms that are strictly pathogens (e.g, Neisseria gonorrhoeae, rabies virus). Perhaps it is not surprising that a general understanding of the microbiota is essential in the evaluation of infectious diseases. Individuals’ microbiotas likely have a major impact on their susceptibility to infectious diseases and even their responses to vaccines. Site-specific knowledge of the indigenous flora may facilitate appropriate interpretation of culture results, aid in selection of empirical antimicrobial therapy based on the likely causative agents, and provide additional impetus for rational antibiotic use to minimize the untoward effects of these drugs on the “beneficial” microbes that inhabit the body.
WHEN TO CONSIDER AN INFECTIOUS ETIOLOGY
The title of this chapter may appear to presuppose that the physician knows when a patient has an infectious disease. In reality, this chapter can serve only as a guide to the evaluation of a patient in whom an infectious disease is a possibility. Once a specific diagnosis is made, the reader should consult the subsequent chapters that deal with specific microorganisms in detail. The challenge for the physician is to recognize which patients may have an infectious disease as opposed to some other underlying disorder. This task is greatly complicated by the fact that infections have an infinite range of presentations, from acute life-threatening conditions (e.g., meningococcemia) to chronic diseases of varying severity (e.g., H. pylori–associated peptic ulcer disease) to no symptoms at all (e.g., latent M. tuberculosis infection). While it is impossible to generalize about a presentation that encompasses all infections, common findings in the history, physical examination, and basic laboratory testing often suggest that the patient either has an infectious disease or should be more closely evaluated for one. This chapter focuses on these common findings and how they may direct the ongoing evaluation of the patient.
PERSPECTIVE
The study of infectious diseases is really a study of host-bacterial interactions and represents evolution by both the host and the bacteria—an endless struggle in which microbes have generally been more creative and adaptive. Given that nearly one-quarter of deaths worldwide are still related to infectious diseases, it is clear that the war against infectious diseases has not been won. For example, a cure for HIV infection is still lacking, there have been only marginal improvements in the methods for detection and treatment of tuberculosis after more than a half century of research, new infectious diseases (e.g., pandemic influenza, viral hemorrhagic fevers) continue to emerge, and the threat of microbial bioterrorism remains high. The subsequent chapters in Part 8 detail—on both a syndrome and a microbe-by-microbe basis—the current state of medical knowledge about infectious diseases. At their core, all of these chapters carry a similar message: Despite numerous advances in the diagnosis, treatment, and prevention of infectious diseases, much work and research are required before anyone can confidently claim that “all the major infections have disappeared.” In reality, this goal will never be attained, given the rapid adaptability of microbes.
145e |
Molecular Mechanisms of Microbial Pathogenesis |
Over the past four decades, molecular studies of the pathogenesis of microorganisms have yielded an explosion of information about the various microbial and host molecules that contribute to the processes of infection and disease. These processes can be classified into several stages: microbial encounter with and entry into the host; microbial growth after entry; avoidance of innate host defenses; tissue invasion and tropism; tissue damage; and transmission to new hosts. Virulence is the measure of an organism’s capacity to cause disease and is a function of the pathogenic factors elaborated by microbes. These factors promote colonization (the simple presence of potentially pathogenic microbes in or on a host), infection (attachment and growth of pathogens and avoidance of host defenses), and disease (often, but not always, the result of activities of secreted toxins or toxic metabolites). In addition, the host’s inflammatory response to infection greatly contributes to disease and its attendant clinical signs and symptoms. The recent surge of interest in the role of the microbiota and its associated microbiome—the collection of microbial genomes residing in or on mammalian organisms—in the physiology of, susceptibility to, and response to infection and in immune system development has had an enormous impact on our understanding of host-pathogen interaction.
THE MICROBIOME
(See also Chap. 86e) We now understand that the indigenous microbial organisms living in close association with almost all animals are organized into complex communities that strongly modulate the ability of pathogenic microbes to become established in or on host surfaces. The sheer numbers of these microbes and their genomic variability vastly exceed the numbers of host cells and genes in a typical animal. Changes and differences in microbiomes within and between individuals, currently characterized by high-throughput DNA sequencing techniques and bioinformatic analysis, affect the development and control of the immune system as well as such diverse conditions as obesity, type 1 diabetes, cognition, neurologic states, autoimmune diseases, and infectious diseases of the skin, gastrointestinal tract, respiratory tract, and vagina. It has been more difficult to directly associate specific types of microbiomes with pathophysiologic states and to assess how conserved or variable microbial species within human and animal microbiomes are evolving. Defining clusters of organisms associated with diseases may become more feasible as more data are obtained. Complicating this task are the results from the Human Microbiome Project suggesting a high level of variability among individuals in the components of the microbiome, although many individuals appear to maintain a fairly conserved microbiome throughout their lives. In the context of infectious diseases, clear changes and disruptions of the indigenous microbiome have a strong and often fundamental impact on the progression of infection. Such alterations can be associated with the effects of antibiotic and immunosuppressive drug use on the normal flora, with environmental changes, and with the impact of microbial virulence factors that displace the indigenous microbial flora to facilitate pathogen colonization. As the available technology for defining the microbiome expands, there is no doubt that the resulting data will markedly affect our concepts of and approaches to microbial pathogenesis and infectious disease treatment.
MICROBIAL ENTRY AND ADHERENCE
Entry Sites A microbial pathogen can potentially enter any part of a host organism. In general, the type of disease produced by a particular microbe is often a direct consequence of its route of entry into the body. The most common sites of entry are mucosal surfaces (the respiratory, alimentary, and urogenital tracts) and the skin. Ingestion, inhalation, and sexual contact are typical routes of microbial entry. Other portals of entry include sites of skin injury (cuts, bites, burns, trauma) along with injection via natural (i.e., vector-borne) or artificial (i.e., needle-stick injury) routes. A few pathogens, such as Schistosoma species, can penetrate unbroken skin. The conjunctiva can serve as an entry point for pathogens of the eye, which occasionally spread systemically from that site.
Microbial entry usually relies on the presence of specific factors needed for persistence and growth in a tissue. Fecal-oral spread via the alimentary tract requires a biologic profile consistent with survival in the varied environments of the gastrointestinal tract (including the low pH of the stomach and the high bile content of the intestine) as well as in contaminated food or water outside the host. Organisms that gain entry via the respiratory tract survive well in small moist droplets produced during sneezing and coughing. Pathogens that enter by venereal routes often survive best in the warm moist environment of the urogenital mucosa and have restricted host ranges (e.g., Neisseria gonorrhoeae, Treponema pallidum, and HIV).
The biology of microbes entering through the skin is highly varied. Some of these organisms can survive in a broad range of environments, such as the salivary glands or alimentary tracts of arthropod vectors, the mouths of larger animals, soil, and water. A complex biology allows protozoan parasites such as Plasmodium, Leishmania, and Trypanosoma species to undergo morphogenic changes that permit transmission to mammalian hosts during insect feeding for blood meals. Plasmodia are injected as infective sporozoites from the salivary glands during mosquito feeding. Leishmania parasites are regurgitated as promastigotes from the alimentary tract of sandflies and injected by bite into a susceptible host. Trypanosomes are first ingested from infected hosts by reduviid bugs; the pathogens then multiply in the gastrointestinal tract of the insects and are released in feces onto the host’s skin during subsequent feedings. Most microbes that land directly on intact skin are destined to die, as survival on the skin or in hair follicles requires resistance to fatty acids, low pH, and other antimicrobial factors on the skin. Once it is damaged (and particularly if it becomes necrotic), the skin can be a major portal of entry and growth for pathogens and elaboration of their toxic products. Burn wound infections and tetanus are clear examples. After animal bites, pathogens resident in the animal’s saliva gain access to the victim’s tissues through the damaged skin. Rabies is the paradigm for this pathogenic process; rabies virus grows in striated muscle cells at the site of inoculation.
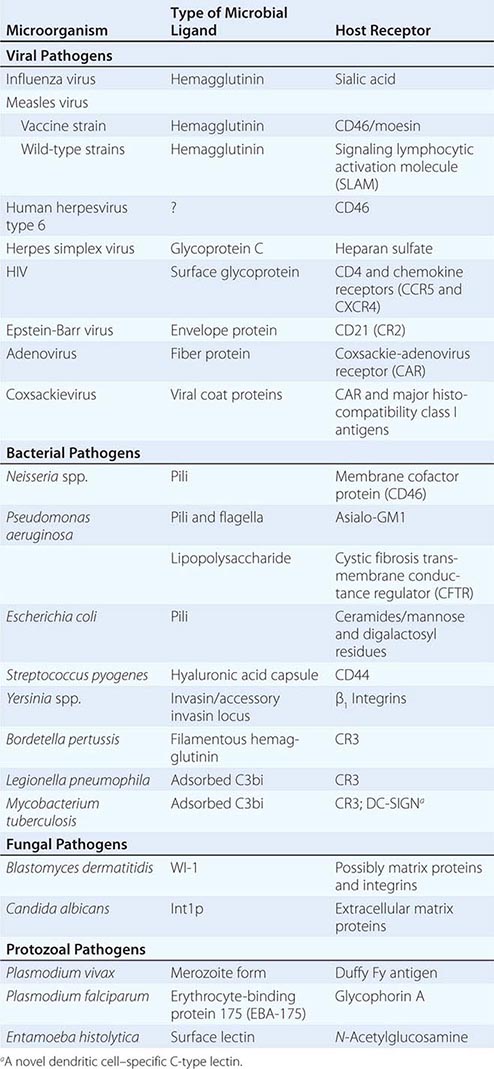
Microbial Adherence Once in or on a host, most microbes must anchor themselves to a tissue or tissue factor; the possible exceptions are organisms that directly enter the bloodstream and multiply there. Specific ligands or adhesins for host receptors constitute a major area of study in the field of microbial pathogenesis. Adhesins comprise a wide range of surface structures, not only anchoring the microbe to a tissue and promoting cellular entry where appropriate but also eliciting host responses critical to the pathogenic process (Table 145e-1). Most microbes produce multiple adhesins specific for multiple host receptors. These adhesins are often redundant, are serologically variable, and act additively or synergistically with other microbial factors to promote microbial sticking to host tissues. In addition, some microbes adsorb host proteins onto their surface and utilize the natural host protein receptor for microbial binding and entry into target cells.
|
EXAMPLES OF MICROBIAL LIGAND-RECEPTOR INTERACTIONS |
VIRAL ADHESINS All viral pathogens must bind to host cells, enter them, and replicate within them. Viral coat proteins serve as the ligands for cellular entry, and more than one ligand-receptor interaction may be needed; for example, HIV utilizes its envelope glycoprotein (gp) 120 to enter host cells by binding both to CD4 and to one of two receptors for chemokines (designated CCR5 and CXCR4). Similarly, the measles virus H glycoprotein binds to both CD46 and the membrane-organizing protein moesin on host cells. The gB and gC proteins on herpes simplex virus bind to heparan sulfate, although this adherence is not essential for entry but rather serves to concentrate virions close to the cell surface; this step is followed by attachment to mammalian cells mediated by the viral gD protein, with subsequent formation of a homotrimer of viral gB protein or a heterodimer of viral gH and gL proteins that permits fusion of the viral envelope with the host cell membrane. Herpes simplex virus can use a number of eukaryotic cell surface receptors for entry, including the herpesvirus entry mediator (related to the tumor necrosis factor receptor), members of the immunoglobulin superfamily, the proteins nectin-1 and nectin-2, and modified heparan sulfate.
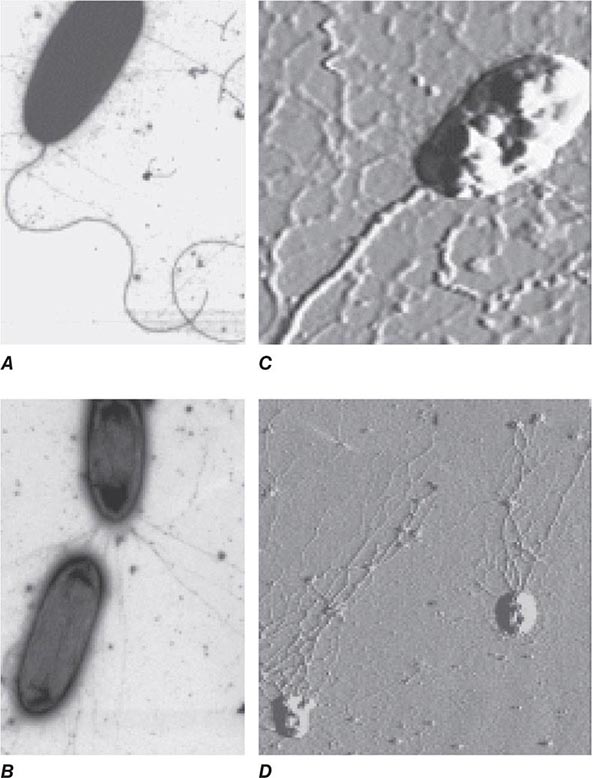
BACTERIAL ADHESINS Among the microbial adhesins studied in greatest detail are bacterial pili and flagella (Fig. 145e-1). Pili or fimbriae are commonly used by gram-negative bacteria for attachment to host cells and tissues; studies have identified similar factors produced by gram-positive organisms such as group B streptococci. In electron micrographs, these hairlike projections (up to several hundred per cell) may be confined to one end of the organism (polar pili) or distributed more evenly over the surface. An individual cell may have pili with a variety of functions. Most pili are made up of a major pilin protein subunit (molecular weight, 17,000–30,000) that polymerizes to form the pilus. Many strains of Escherichia coli isolated from urinary tract infections express mannose-binding type 1 pili, whose binding to integral membrane glycoproteins called uroplakins that coat the cells in the bladder epithelium is inhibited by D-mannose. Other strains produce the Pap (pyelonephritis-associated) or P pilus adhesin that mediates binding to digalactose (gal-gal) residues on globosides of the human P blood groups. Both of these types of pili have proteins located at the tips of the main pilus unit that are critical to the binding specificity of the whole pilus unit. Although immunization with the mannose-binding tip protein (FimH) of type 1 pili prevents experimental E. coli bladder infections in mice and monkeys, a human trial of this vaccine was not successful. E. coli cells causing diarrheal disease express pilus-like receptors for enterocytes on the small bowel, along with other receptors termed colonization factors.
FIGURE 145e-1 Bacterial surface structures. A and B. Traditional electron micrographic images of fixed cells of Pseudomonas aeruginosa. Flagella (A) and pili (B) project out from the bacterial poles. C and D. Atomic force microscopic image of live P. aeruginosa freshly planted onto a smooth mica surface. This technology reveals the fine, three-dimensional detail of the bacterial surface structures. (Images courtesy of Drs. Martin Lee and Milan Bajmoczi, Harvard Medical School.)
The type IV pilus, a common type of pilus found in Neisseria species, Moraxella species, Vibrio cholerae, Legionella pneumophila, Salmonella enterica serovar Typhi, enteropathogenic E. coli, and Pseudomonas aeruginosa, often mediates adherence of organisms to target surfaces. Type IV pili tend to have a relatively conserved aminoterminal region and a more variable carboxyl-terminal region. For some species (e.g., N. gonorrhoeae, Neisseria meningitidis, and enteropathogenic E. coli), the pili are critical for attachment to mucosal epithelial cells. For others, such as P. aeruginosa, the pili only partially mediate the cells’ adherence to host tissues and may in some circumstances inhibit colonization. For example, a recent study of P. aeruginosa colonization of the gastrointestinal tract of mice evaluated a bank of mutants in which all nonessential genes were interrupted; those mutants that were unable to produce the type IVa pili were actually better able to colonize the gastrointestinal mucosa, although the basis for this observation was not identified. V. cholerae cells appear to use two different types of pili for intestinal colonization. Whereas interference with this stage of colonization would appear to be an effective antibacterial strategy, attempts to develop pilus-based vaccines for human diseases have not been highly successful to date.
Flagella are long appendages attached at either one or both ends of the bacterial cell (polar flagella) or distributed over the entire cell surface (peritrichous flagella). Flagella, like pili, are composed of a polymerized or aggregated basic protein. In flagella, the protein subunits form a tight helical structure and vary serologically with the species. Spirochetes such as T. pallidum and Borrelia burgdorferi have axial filaments similar to flagella running down the long axis of the center of the cell, and they “swim” by rotation around these filaments. Some bacteria can glide over a surface in the absence of obvious motility structures.
Other bacterial structures involved in adherence to host tissues include specific staphylococcal and streptococcal proteins that bind to human extracellular matrix proteins such as fibrin, fibronectin, fibrinogen, laminin, and collagen. Fibronectin appears to be a commonly used receptor for various pathogens; a particular amino acid sequence in fibronectin, Arg-Gly-Asp or RGD, is a critical target used by bacteria to bind to host tissues. Binding of a highly conserved Staphylococcus aureus surface protein, clumping factor A (ClfA), to fibrinogen has been implicated in many aspects of pathogenesis. Attempts to interrupt this interaction and prevent S. aureus sepsis in low-birth-weight infants by administering an intravenous IgG preparation derived from the plasma of individuals with high titers of antibody to ClfA failed to show efficacy in a clinical trial; however, this approach is being pursued in some vaccine formulations targeting this organism. The conserved outer-core portion of the lipopolysaccharide (LPS) of P. aeruginosa mediates binding to the cystic fibrosis transmembrane conductance regulator (CFTR) on airway epithelial cells—an event that appears to play a critical role in normal host resistance to infection by initiating recruitment of polymorphonuclear neutrophils (PMNs) to the lung mucosa to kill the cells via opsonophagocytosis. A large number of microbial pathogens encompassing major gram-positive bacteria (staphylococci and streptococci), gram-negative bacteria (major enteric species and coccobacilli), fungi (Candida, Fusobacterium, Aspergillus), and even eukaryotes (Trichomonas vaginalis and Plasmodium falciparum) express a surface polysaccharide composed of β-1-6-linked-poly-N-acetyl-D-glucosamine (PNAG). One of the functions of PNAG for some of these organisms is to promote binding to materials used in catheters and other types of implanted devices. This polysaccharide may be a critical factor in the establishment of device-related infections by pathogens such as staphylococci and E. coli. High-powered imaging techniques (e.g., atomic force microscopy) have revealed that bacterial cells have a nonhomogeneous surface that is probably attributable to different concentrations of cell surface molecules, including microbial adhesins, at specific places on the cell surface (Figs. 120-1C and 120-1D).
FUNGAL ADHESINS Several fungal adhesins have been described that mediate colonization of epithelial surfaces, particularly adherence to structures like fibronectin, laminin, and collagen. The product of the Candida albicans INT1 gene, Int1p, bears similarity to mammalian integrins that bind to extracellular matrix proteins. The agglutinin-like sequence (ALS) adhesins are large cell-surface glycoproteins mediating adherence of pathogenic Candida to host tissues. These adhesins possess a conserved three-domain structure composed of an N-terminal domain that mediates adherence to host tissue receptors, a central motif consisting of a number of repeats of a conserved sequence of 36 amino acids, and a C-terminal domain that varies in length and sequence and contains a glycosylphosphatidylinositol (GPI) anchor addition site that allows binding of the adhesin to the fungal cell wall. Variability in the number of central domains in different ALS proteins characterizes different adhesins with specificity for different host receptors. The ALS adhesins are expressed under certain environmental conditions and are crucial for pathogenesis of fungal infections.
For several fungal pathogens that initiate infections after inhalation of infectious material, the inoculum is ingested by alveolar macrophages, in which the fungal cells transform to pathogenic phenotypes. Like C. albicans, Blastomyces dermatitidis binds to CD11b/CD18 integrins as well as to CD14 on macrophages. B. dermatitidis produces a 120-kDa surface protein, designated WI-1, that mediates this adherence. An unidentified factor on Histoplasma capsulatum also mediates binding of this fungal pathogen to the integrin surface proteins.
EUKARYOTIC PATHOGEN ADHESINS Eukaryotic parasites use complicated surface glycoproteins as adhesins, some of which are lectins (proteins that bind to specific carbohydrates on host cells). For example, Plasmodium vivax, one of six Plasmodium species causing malaria, binds (via Duffy-binding protein) to the Duffy blood group carbohydrate antigen Fy on erythrocytes. Entamoeba histolytica, the third leading cause of death from parasitic diseases, expresses two proteins that bind to the disaccharide galactose/N-acetyl galactosamine. Reports indicate that children with mucosal IgA antibody to one of these lectins are resistant to reinfection with virulent E. histolytica. A major surface glycoprotein (gp63) of Leishmania promastigotes is needed for these parasites to enter human macrophages—the principal target cell of infection. This glycoprotein promotes complement binding but inhibits complement lytic activity, allowing the parasite to use complement receptors for entry into macrophages; gp63 also binds to fibronectin receptors on macrophages. In addition, the pathogen can express a carbohydrate that mediates binding to host cells. Evidence suggests that, as part of hepatic granuloma formation, Schistosoma mansoni expresses a carbohydrate epitope related to the Lewis × blood group antigen that promotes adherence of helminthic eggs to vascular endothelial cells under inflammatory conditions.
Host Receptors Host receptors are found both on target cells (such as epithelial cells lining mucosal surfaces) and within the mucus layer covering these cells. Microbial pathogens bind to a wide range of host receptors to establish infection (Table 145e-1). Selective loss of host receptors for a pathogen may confer natural resistance to an otherwise susceptible population. For example, 70% of individuals in West Africa lack Fy antigens and are resistant to P. vivax infection. S. enterica serovar Typhi, the etiologic agent of typhoid fever, produces a pilus protein that binds to CFTR to enter the gastrointestinal submucosa after being ingested by enterocytes. As homozygous mutations in CFTR are the cause of the life-shortening disease cystic fibrosis, heterozygote carriers (e.g., 4–5% of individuals of European ancestry) may have had a selective advantage due to decreased susceptibility to typhoid fever.
Numerous virus–target cell interactions have been described, and it is now clear that different viruses can use similar host cell receptors for entry. The list of certain and likely host receptors for viral pathogens is long. Among the host membrane components that can serve as receptors for viruses are sialic acids, gangliosides, glycosaminoglycans, integrins and other members of the immunoglobulin superfamily, histocompatibility antigens, and regulators and receptors for complement components. A notable example of the effect of host receptors on the pathogenesis of infection has emerged from studies comparing the binding of avian influenza A subtype H5N1 with that of influenza A strains expressing the H1 subtype of hemagglutinin. The H1 subtypes tend to be highly pathogenic and transmissible from human to human, and they bind to a receptor composed of two sugar molecules: sialic acid linked α-2-6 to galactose. This receptor is expressed at high levels in the airway epithelium; when virus is shed from this surface, its transmission via coughing and aerosol droplets is facilitated. In contrast, the H5N1 avian influenza virus binds to sialic acid linked α-2-3 to galactose, and this receptor is expressed at high levels in pneumocytes in the alveoli. Infection in the alveoli is thought to underlie the high mortality rate associated with avian influenza but also the low interhuman transmissibility of this strain, which is not readily transported to the airways from which it can be expelled by coughing. Nonetheless, it was recently shown that H5 hemagglutinins can acquire mutations that vastly increase their transmissibility while not affecting their high level of lethality.
MICROBIAL GROWTH AFTER ENTRY
Once established on a mucosal or skin site, pathogenic microbes must replicate before causing full-blown infection and disease. Within cells, viral particles release their nucleic acids, which may be directly translated into viral proteins (positive-strand RNA viruses), transcribed from a negative strand of RNA into a complementary mRNA (negative-strand RNA viruses), or transcribed into a complementary strand of DNA (retroviruses); for DNA viruses, mRNA may be transcribed directly from viral DNA, either in the cell nucleus or in the cytoplasm. To grow, bacteria must acquire specific nutrients or synthesize them from precursors in host tissues. Many infectious processes are usually confined to specific epithelial surfaces—e.g., H1 subtype influenza to the respiratory mucosa, gonorrhea to the urogenital epithelium, shigellosis to the gastrointestinal epithelium. While there are multiple reasons for this specificity, one important consideration is the ability of these pathogens to obtain from these specific environments the nutrients needed for growth and survival.
Temperature restrictions also play a role in limiting certain pathogens to specific tissues. Rhinoviruses, a cause of the common cold, grow best at 33°C and replicate in cooler nasal tissues but not in the lung. Leprosy lesions due to Mycobacterium leprae are found in and on relatively cool body sites. Fungal pathogens that infect the skin, hair follicles, and nails (dermatophyte infections) remain confined to the cooler, exterior, keratinous layer of the epithelium.
A topic of major interest is the ability of many bacterial, fungal, and protozoal species to grow in multicellular masses referred to as biofilms. These masses are biochemically and morphologically quite distinct from the free-living individual cells referred to as planktonic cells. Growth in biofilms leads to altered microbial metabolism, production of extracellular virulence factors, and decreased susceptibility to biocides, antimicrobial agents, and host defense molecules and cells. P. aeruginosa growing on the bronchial mucosa during chronic infection, staphylococci and other pathogens growing on implanted medical devices, and dental pathogens growing on tooth surfaces to form plaque are several examples of microbial biofilm growth associated with human disease. Many other pathogens can form biofilms during in vitro growth. It is increasingly accepted that this mode of growth contributes to microbial virulence and induction of disease and that biofilm formation can also be an important factor in microbial survival outside the host, promoting transmission to additional susceptible individuals.
AVOIDANCE OF INNATE HOST DEFENSES
As microbes have interacted with mucosal/epithelial surfaces since the emergence of multicellular organisms, it is not surprising that multicellular hosts have a variety of innate surface defense mechanisms that can sense when pathogens are present and contribute to their elimination. The skin is acidic and is bathed with fatty acids toxic to many microbes. Skin pathogens such as staphylococci must tolerate these adverse conditions. Mucosal surfaces are covered by a barrier composed of a thick mucus layer that entraps microbes and facilitates their transport out of the body by such processes as mucociliary clearance, coughing, and urination. Mucous secretions, saliva, and tears contain antibacterial factors such as lysozyme and antimicrobial peptides as well as antiviral factors such as interferons (IFNs). Gastric acidity and bile salts are inimical to the survival of many ingested pathogens, and most mucosal surfaces—particularly the nasopharynx, the vaginal tract, and the gastrointestinal tract—contain a resident flora of commensal microbes that interfere with the ability of pathogens to colonize and infect a host. Major advances in the use of nucleic acid sequencing now allow extensive identification and characterization of the vast array of commensal organisms that have come to be referred to as the microbiota. In addition to its role in providing competition for mucosal colonization, acquisition of a normal microbiota is critical for proper development of the immune system, influencing maturation and differentiation of components of both the innate and acquired arms.
Pathogens that survive local antimicrobial factors must still contend with host endocytic, phagocytic, and inflammatory responses as well as with host genetic factors that determine the degree to which a pathogen can survive and grow. The list of genes whose variants, usually by single-nucleotide polymorphisms, can affect host susceptibility and resistance to infection is rapidly expanding. A classic example is a 32-bp deletion in the gene for the HIV-1 co-receptor known as chemokine receptor 5 (CCR5), which, when present in the homozygous state, confers high-level resistance to HIV-1 infection. The growth of viral pathogens entering skin or mucosal epithelial cells can be limited by a variety of host genetic factors, including production of IFNs, modulation of receptors for viral entry, and age- and hormone-related susceptibility factors; by nutritional status; and even by personal habits such as smoking and exercise.
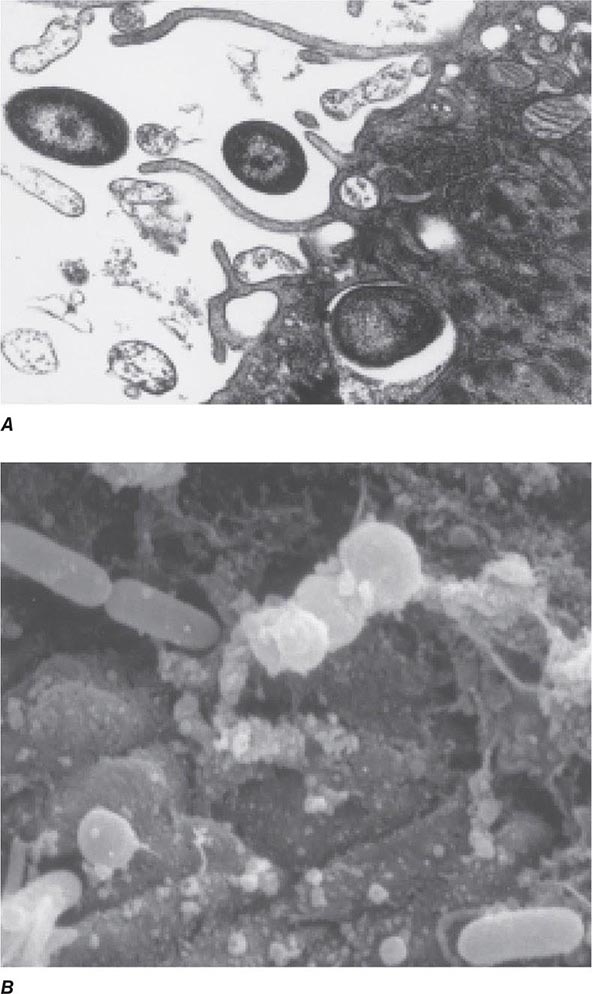
Encounters with Epithelial Cells Over the past two decades, many pathogens have been shown to enter epithelial cells (Fig. 145e-2); they often use specialized surface structures that bind to receptors, with consequent internalization. However, the exact role and the importance of this process in infection and disease are not well defined for most of these pathogens. Microbial entry into host epithelial cells is seen as a means for dissemination to adjacent or deeper tissues or as a route to sanctuary to avoid ingestion and killing by professional phagocytes. Epithelial cell entry appears, for instance, to be a critical aspect of dysentery induction by Shigella.
FIGURE 145e-2 Entry of bacteria into epithelial cells. A. Internalization of Pseudomonas aeruginosa by cultured airway epithelial cells expressing wild-type cystic fibrosis transmembrane conductance regulator, the cell receptor for bacterial ingestion. B. Entry of P. aeruginosa into murine tracheal epithelial cells after murine infection by the intranasal route.
Curiously, the less virulent strains of many bacterial pathogens are more adept at entering epithelial cells than are more virulent strains; examples include pathogens that lack the surface polysaccharide capsule needed to cause serious disease. Thus, for Haemophilus influenzae, Streptococcus pneumoniae, Streptococcus agalactiae (group B Streptococcus), and Streptococcus pyogenes, isogenic mutants or variants lacking capsules enter epithelial cells better than the wild-type, encapsulated parental forms that cause disseminated disease. These observations have led to the proposal that epithelial cell entry may be primarily a manifestation of host defense, resulting in bacterial clearance by both shedding of epithelial cells containing internalized bacteria and initiation of a protective and nonpathogenic inflammatory response. However, a possible consequence of this process could be the opening of a hole in the epithelium, potentially allowing uningested organisms to enter the submucosa. This scenario has been documented in murine S. enterica serovar Typhimurium infections and in experimental bladder infections with uropathogenic E. coli. In the latter system, bacterial pilus-mediated attachment to uroplakins induces exfoliation of the cells with attached bacteria. Subsequently, infection is produced by residual bacterial cells that invade the superficial bladder epithelium, where they can grow intracellularly into biofilm-like masses encased in an extracellular polysaccharide-rich matrix and surrounded by uroplakin. This mode of growth produces structures that have been referred to as bacterial pods. It is likely that at low bacterial inocula epithelial cell ingestion and subclinical inflammation are efficient means to eliminate pathogens, while at higher inocula a proportion of surviving bacterial cells enter the host tissue through the damaged mucosal surface and multiply, producing disease. Alternatively, failure of the appropriate epithelial cell response to a pathogen may allow the organism to survive on a mucosal surface where, if it avoids other host defenses, it can grow and cause a local infection. Along these lines, as noted above, P. aeruginosa is taken into epithelial cells by CFTR, a protein missing or nonfunctional in most severe cases of cystic fibrosis. The major clinical consequence of this disease is chronic airway-surface infection with P. aeruginosa in 80–90% of patients. The failure of airway epithelial cells to ingest and promote the removal of P. aeruginosa via a properly regulated inflammatory response has been proposed as a key component of the hypersusceptibility of cystic fibrosis patients to chronic airway infection with this organism.
Encounters with Phagocytes • PHAGOCYTOSIS AND INFLAMMATION Phagocytosis of microbes is a major innate host defense that limits the growth and spread of pathogens. Phagocytes appear rapidly at sites of infection in conjunction with the initiation of inflammation. Ingestion of microbes by both tissue-fixed macrophages and migrating phagocytes probably accounts for the limited ability of most microbial agents to cause disease. A family of related molecules called collectins, soluble defense collagens, or pattern-recognition molecules are found in blood (mannose-binding lectins), in lung (surfactant proteins A and D), and most likely in other tissues as well and bind to carbohydrates on microbial surfaces to promote phagocyte clearance. Bacterial pathogens seem to be ingested principally by PMNs, while eosinophils are frequently found at sites of infection by protozoan or multicellular parasites. Successful pathogens, by definition, must avoid being cleared by professional phagocytes. One of several antiphagocytic strategies employed by bacteria and by the fungal pathogen Cryptococcus neoformans is to elaborate large-molecular-weight surface polysaccharide antigens, often in the form of a capsule that coats the cell surface. Most pathogenic bacteria produce such antiphagocytic capsules. On occasion, proteins or polypeptides form capsule-like coatings for organisms such as group A streptococci and Bacillus anthracis.
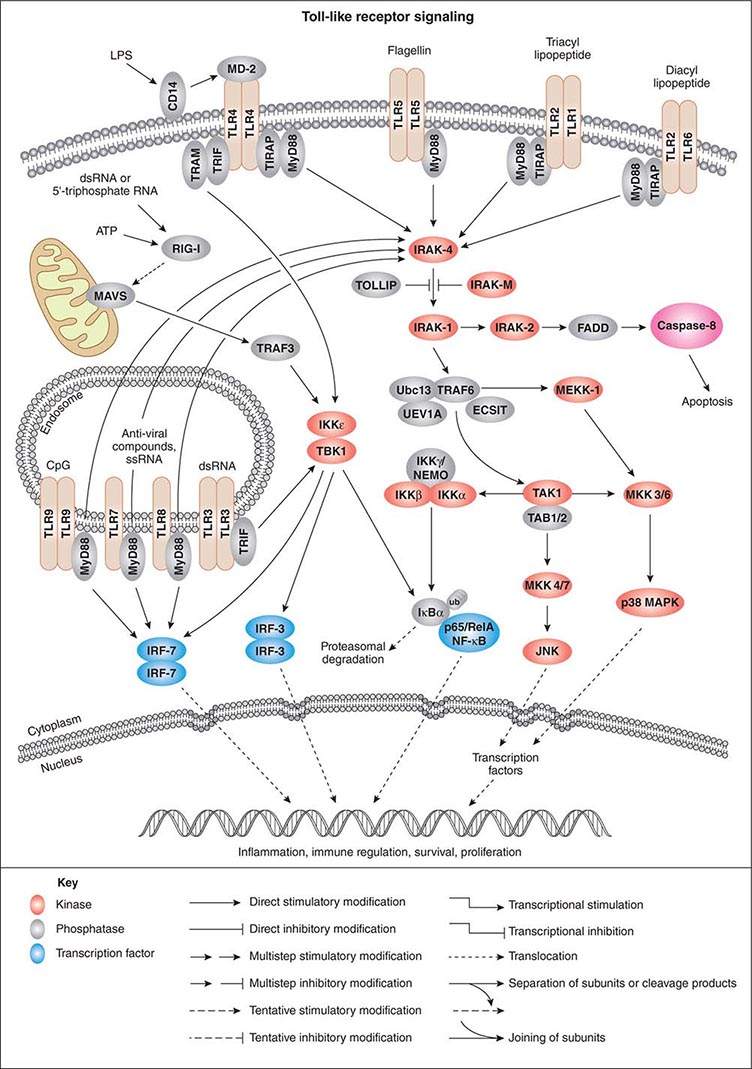
As activation of local phagocytes in tissues is a key step in initiating inflammation and migration of additional phagocytes into infected sites, much attention has been paid to microbial factors that initiate inflammation. These are usually conserved factors critical to the microbes’ survival and are referred to as pathogen-associated molecular patterns (PAMPs). Cellular responses to microbial encounters with phagocytes are governed largely by the structure of the microbial PAMPs that elicit inflammation, and detailed knowledge of these structures of bacterial pathogens has contributed greatly to our understanding of molecular mechanisms of microbial pathogenesis mediated by activation of host cell molecules such as TLRs (Fig. 145e-3). One of the best-studied systems involves the interaction of LPS from gram-negative bacteria and the GPI-anchored membrane protein CD14 found on the surface of professional phagocytes, including migrating and tissue-fixed macrophages and PMNs. A soluble form of CD14 is also found in plasma and on mucosal surfaces. A plasma protein, LPS-binding protein, transfers LPS to membrane-bound CD14 on myeloid cells and promotes binding of LPS to soluble CD14. Soluble CD14/LPS/LPS-binding protein complexes bind to many cell types and may be internalized to initiate cellular responses to microbial pathogens. It has been shown that peptidoglycan and lipoteichoic acid from gram-positive bacteria as well as cell-surface products of mycobacteria and spirochetes can interact with CD14 (Fig. 145e-3). Additional molecules, such as MD-2, also participate in the recognition of bacterial activators of inflammation.
FIGURE 145e-3 Cellular signaling pathways for production of inflammatory cytokines in response to microbial products. Microbial cell-surface constituents interact with Toll-like receptors (TLRs), in some cases requiring additional factors such as MD-2, which facilitates the response to lipopolysaccharide (LPS) via TLR4. Although these constituents are depicted as interacting with the TLRs on the cell surface, TLRs contain extracellular leucine-rich domains that become localized to the lumen of the phagosome upon uptake of bacterial cells. The internalized TLRs can bind to microbial products. The TLRs are oligomerized, usually forming homodimers, and then bind to the general adapter protein MyD88 via the C-terminal Toll/IL-1R (TIR) domains, which also bind to TIRAP (TIR domain-containing adapter protein), a molecule that participates in the transduction of signals from TLRs 1, 2, 4, and 6. The MyD88/TIRAP complex activates signal-transducing molecules such as IRAK-4 (IL-1Rc-associated kinase 4), which in turn activates IRAK-1. This activation can be blocked by IRAK-M and Toll-interacting protein (TOLLIP). IRAK-1 activates TRAF6 (tumor necrosis factor receptor–associated factor 6), TAK1 (transforming growth factor β–activating kinase 1), and TAB1/2 (TAK1-binding protein 1/2). This signaling complex associates with the ubiquitin-conjugating enzyme Ubc13 and the Ubc-like protein UEV1A to catalyze the formation of a polyubiquitin chain on TRAF6. Polyubiquitination of TRAF6 activates TAK1, which, along with TAB1/2 (a protein that binds to lysine residue 63 in polyubiquitin chains via a conserved zinc-finger domain), phosphorylates the inducible kinase complex: IKKα, IKKβ, and IKKγ. IKKγ is also called NEMO (nuclear factor κB [NF-κB] essential modulator). This large complex phosphorylates the inhibitory component of NF-κB, IκBα, resulting in release of IκBα from NF-κB. Phosphorylated (PP) IκB is then ubiquitinated (ub) and degraded, and the two components of NF-κB, p50 or Rel and p65, translocate to the nucleus, where they bind to regulatory transcriptional sites on target genes, many of which encode inflammatory proteins. In addition to inducing NF-κB nuclear translocation, the TAK1/TAB1/2 complex activates MAP kinase transducers such as MKK 4/7 and MKK 3/6, which can lead to nuclear translocation of transcription factors such as AP1. TLR4 can also activate NF-κB nuclear translocation via the MyD88-independent TRIF (TIR domain–containing adapter-inducing IFN-β) and TRAM (TRIF-related adapter molecule) cofactors. Intracellular TLRs 3, 7, 8, and 9 also use MyD88 and TRIF to activate IFN response factors 3 and 7 (IRF-3 and IRF-7), which also function as transcriptional factors in the nucleus. ATP, adenosine 5’-triphosphate; ECSIT, evolutionarily conserved signaling intermediate in Toll pathways; FADD, Fas-associated protein with death domain; JNK, c-Jun N-terminal kinase; MAVS, mitochondrial antiviral signaling protein; MEKK-1, MAP/ERK kinase kinase 1; p38 MAPK, p38 mitogen-activated protein kinase; RIG-1, retinoic acid–inducible gene 1; TBK1, TANK-binding kinase 1. (Pathway diagram reproduced courtesy of Cell Signaling Technology, Inc. [www.cellsignal.com].)
GPI-anchored receptors do not have intracellular signaling domains; therefore, it is the TLRs that transduce signals for cellular activation due to LPS binding. Binding of microbial factors to TLRs to activate signal transduction occurs in the phagosome—and not on the surface—of dendritic cells that have internalized the microbe. This binding is probably due to the release of the microbial surface factor from the cell in the environment of the phagosome, where the liberated factor can bind to its cognate TLRs. TLRs initiate cellular activation through a series of signal-transducing molecules (Fig. 145e-3) that lead to nuclear translocation of the transcription factor NF-κB (nuclear factor κB), a master-switch for production of important inflammatory cytokines such as tumor necrosis factor α (TNF-α) and interleukin (IL) 1.
The initiation of inflammation can occur not only with LPS and peptidoglycan but also with viral particles and other microbial products such as polysaccharides, enzymes, and toxins. Bacterial flagella activate inflammation by binding of a conserved sequence to TLR5. Some pathogens (e.g., Campylobacter jejuni, Helicobacter pylori, and Bartonella bacilliformis) make flagella that lack this sequence and do not bind to TLR5; thus efficient host responses to infection are prevented. Bacteria also produce a high proportion of DNA molecules with unmethylated CpG residues that activate inflammation through TLR9. TLR3 recognizes double-stranded RNA, a pattern-recognition molecule produced by many viruses during their replicative cycle. TLR1 and TLR6 associate with TLR2 to promote recognition of acylated microbial proteins and peptides.
The myeloid differentiation factor 88 (MyD88) molecule and the Toll/IL-1R (TIR) domain-containing adapter protein (TIRAP) bind to the cytoplasmic domains of TLRs and also to receptors that are part of the IL-1 receptor families. Numerous studies have shown that MyD88/TIRAP-mediated transduction of signals from TLRs and other receptors is critical for innate resistance to infection, activating MAP-kinases and NF-κB and thereby leading to production of cytokines/chemokines. Mice lacking MyD88 are more susceptible than normal mice to infections with a broad range of pathogens. In one study, nine children homozygous for defective MyD88 genes had recurrent infections with S. pneumoniae, S. aureus, and P. aeruginosa—three bacterial species showing increased virulence in MyD88-deficient mice; however, unlike these mice, the MyD88-deficient children seemed to have no greater susceptibility to other bacteria, viruses, fungi, or parasites. Another component of the MyD88-dependent signaling pathway is a molecule known as IL-1 receptor–associated kinase 4 (IRAK-4). Individuals with a homozygous deficiency in genes encoding this protein are at increased risk for S. pneumoniae and S. aureus infections and, to some degree, for P. aeruginosa infections as well.
In addition to their role in MyD88-mediated signaling, some TLRs (e.g., TLR3 and TLR4) can activate signal transduction via a MyD88-independent pathway involving TIR domain–containing, adapter-inducing IFN-β (TRIF) and the TRIF-related adapter molecule (TRAM). Signaling through TRIF and TRAM activates the production of both NF-κB-dependent cytokines/chemokines and type 1 IFNs. The type 1 IFNs bind to the IFN-α receptor composed of two protein chains, IFNAR1 and IFNAR2. Humans produce three type 1 IFNs: IFN-α, IFN-β, and IFN-γ. These molecules activate another class of proteins known as the signal transducer and activator of transcription (STAT) complexes. The STAT factors are important in regulating immune system genes and thus play a critical role in responding to microbial infections.
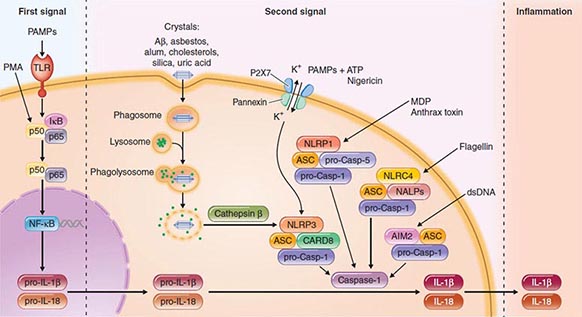
Another intracellular complex of proteins found to be a major factor in the host cell response to infection is the inflammasome (Fig. 145e-4), in which inflammatory cytokines IL-1 and IL-18 are changed from their precursor to their active forms prior to secretion by the cysteine protease caspase-1. Within the inflammasome are additional proteins that are members of the nucleotide binding and oligomerization domain (NOD)–like receptor (NLR) family. Like the TLRs, NOD proteins sense the presence of the conserved microbial factors released inside a cell. Recognition of these PAMPs by NLRs leads to caspase-1 activation and to secretion of active IL-1 and IL-18 by an unknown mechanism. Studies of mice indicate that as many as four inflammasomes with different components are formed: the IPAF inflammasome, the NALP1 inflammasome, the cryopyrin/NALP3 inflammasome, and an inflammasome triggered by Francisella tularensis infection (Fig. 145e-4). The components depend on the type of stimulus driving inflammasome formation and activation.
FIGURE 145e-4 Inflammasomes. The nucleotide-binding oligomerization domain-like receptor (NLR) family of proteins is involved in the regulation of innate immune responses. These proteins sense pathogen-associated molecular patterns (PAMPs) in the cytosol as well as the host-derived signals known as damage-associated molecular patterns (DAMPs). Certain NLRs induce the assembly of large caspase-1-activating complexes called inflammasomes. Activation of caspase-1 through autoproteolytic maturation leads to the processing and secretion of the proinflammatory cytokines interleukin 1β (IL-1β) and IL-18. So far, four inflammasomes have been identified and defined by the NLR protein that they contain: the NLRP1/NALP1b inflammasome; the NLRC4/IPAF inflammasome; the NLRP3/NALP3 inflammasome; and the AIM2 (absent in melanoma 2)–containing inflammasome. Aβ, amyloid β; ASC, apoptosis-associated speck-like protein containing CARD; ATP, adenosine 5’-triphosphate; CARD8, caspase recruitment domain–containing protein 8; IκB, inhibitor of κB; IPAF, interleukin-converting enzyme protease-activating factor; MDP, muramyl dipeptide; NF-κB, nuclear factor κB; P2X7, purinergic P2X7 (receptor); PMA, phorbol myristate acetate; TLR, Toll-like receptor. (Pathway diagram reproduced with permission from Invivogen [www.invivogen.com/review-inflammasome].)
A recent addition to the identified intracellular components responding to microbial infection is autophagy, initially described as an intracellular process for degradation and recycling of cellular components for reuse. Now it is clear that autophagy constitutes an early defense mechanism in which, after ingestion, microbial pathogens either within vacuoles or in the cytoplasm are delivered to lysosomal compartments for degradation. Avoidance of this process is critical if pathogens are to cause disease and can be achieved by multiple mechanisms, such as inhibition of proteins within the autophagic vacuole by shigellae, recruitment of host proteins to mask Listeria monocytogenes, and inhibition of formation of the vacuole by L. pneumophila.
ADDITIONAL INTERACTIONS OF MICROBIAL PATHOGENS AND PHAGOCYTES Other ways that microbial pathogens avoid destruction by phagocytes include production of factors that are toxic to these cells or that interfere with their chemotactic and ingestion function. Hemolysins, leukocidins, and the like are microbial proteins that can kill phagocytes that are attempting to ingest organisms elaborating these substances. For example, S. aureus elaborates a family of bicomponent leukocidins that bind to host receptors such as the HIV co-receptor CCR5 (which is also used by the LukE/D toxin) or—in the case of the Panton-Valentine leukocidin—the receptor of the C5a component of activated complement (which is used by LukF/S). Streptolysin O made by S. pyogenes binds to cholesterol in phagocyte membranes and initiates a process of internal degranulation, with the release of normally granule-sequestered toxic components into the phagocyte’s cytoplasm. E. histolytica, an intestinal protozoan that causes amebic dysentery, can disrupt phagocyte membranes after direct contact via the release of protozoal phospholipase A and pore-forming peptides.
MICROBIAL SURVIVAL INSIDE PHAGOCYTES Many important microbial pathogens use a variety of strategies to survive inside phagocytes (particularly macrophages) after ingestion. Inhibition of fusion of the phagocytic vacuole (the phagosome) containing the ingested microbe with the lysosomal granules containing antimicrobial substances (the lysosome) allows Mycobacterium tuberculosis, S. enterica serovar Typhi, and Toxoplasma gondii to survive inside macrophages. Some organisms, such as L. monocytogenes, escape into the phagocyte’s cytoplasm to grow and eventually spread to other cells. Resistance to killing within the macrophage and subsequent growth are critical to successful infection by herpes-type viruses, measles virus, poxviruses, Salmonella, Yersinia, Legionella, Mycobacterium, Trypanosoma, Nocardia, Histoplasma, Toxoplasma, and Rickettsia. Salmonella species use a master regulatory system—in which the PhoP/PhoQ genes control other genes—to enter and survive within cells, with intracellular survival entailing structural changes in the cell envelope LPS.
TISSUE INVASION AND TISSUE TROPISM
Tissue Invasion Most viral pathogens cause disease by growth at skin or mucosal entry sites, but some pathogens spread from the initial site to deeper tissues. Virus can spread via the nerves (rabies virus) or plasma (picornaviruses) or within migratory blood cells (poliovirus, Epstein-Barr virus, and many others). Specific viral genes determine where and how individual viral strains can spread.
Bacteria may invade deeper layers of mucosal tissue via intracellular uptake by epithelial cells, traversal of epithelial cell junctions, or penetration through denuded epithelial surfaces. Among virulent Shigella strains and invasive strains of E. coli, outer-membrane proteins are critical to epithelial cell invasion and bacterial multiplication. Neisseria and Haemophilus species penetrate mucosal cells by poorly understood mechanisms before dissemination into the bloodstream. Staphylococci and streptococci elaborate a variety of extracellular enzymes, such as hyaluronidase, lipases, nucleases, and hemolysins, that are probably important in breaking down cellular and matrix structures and allowing the bacteria access to deeper tissues and blood. For example, staphylococcal α-hemolysin binds to a receptor, A-disintegrin and metalloprotease 10 (ADAM-10), to cause endothelial cell damage and disruption of vascular barrier function—events that are likely critical for systemic spread of S. aureus from an initial infectious site. Organisms that colonize the gastrointestinal tract can often translocate through the mucosa into the blood and, under circumstances in which host defenses are inadequate, cause bacteremia. Yersinia enterocolitica can invade the mucosa through the activity of the invasin protein. The complex milieu of the basement membrane–containing structures, such as laminin and collagen, that anchor epithelial cells to mucosal surfaces must often be breached. Numerous organisms express factors known as MSCRAMMs (microbial surface components recognizing adhesive matrix molecules). These MSCRAMMS promote bacterial attachment to factors in the host extracellular matrix, such as laminin, collagen, and fibronectin. Additional microbial proteases, along with the host’s own surface-bound plasminogen and host matrix metalloproteases, then combine to degrade the extracellular matrix and promote microbial spread. Some bacteria (e.g., brucellae) can be carried from a mucosal site to a distant site by phagocytic cells that ingest but fail to kill the bacteria.
Fungal pathogens almost always take advantage of host immunocompromise to spread hematogenously to deeper tissues. The AIDS epidemic has resoundingly illustrated this principle: the immunodeficiency of many HIV-infected patients permits the development of life-threatening fungal infections of the lung, blood, and brain. Other than the capsule of C. neoformans, specific fungal antigens involved in tissue invasion are not well characterized. Both fungal pathogens and protozoal pathogens (e.g., Plasmodium species and E. histolytica) undergo morphologic changes to spread within a host. C. albicans undertakes a yeast-hyphal transformation wherein the hyphal forms are found where the fungus is infiltrating the mucosal barrier of tissues, while the yeast form grows on epithelial cell surfaces as well as on the tips of hyphae that have infiltrated tissues. Malarial parasites grow in liver cells as merozoites and are released into the blood to invade erythrocytes and become trophozoites. E. histolytica is found as both a cyst and a trophozoite in the intestinal lumen, through which this pathogen enters the host, but only the trophozoite form can spread systemically to cause amebic liver abscesses. Other protozoal pathogens, such as T. gondii, Giardia lamblia, and Cryptosporidium, also undergo extensive morphologic changes after initial infection to spread to other tissues.
Tissue Tropism The propensity of certain microbes to cause disease by infecting specific tissues has been known since the early days of bacteriology, yet the molecular basis for this propensity is understood somewhat better for viral pathogens than for other agents of infectious disease. Specific receptor-ligand interactions clearly underlie the ability of certain viruses to enter cells within tissues and disrupt normal tissue function, but the mere presence of a receptor for a virus on a target tissue is not sufficient for tissue tropism. Factors in the cell, route of viral entry, viral capacity to penetrate into cells, viral genetic elements that regulate gene expression, and pathways of viral spread in a tissue all affect tissue tropism. Some viral genes are best transcribed in specific target cells, such as hepatitis B genes in liver cells and Epstein-Barr virus genes in B lymphocytes. The route of inoculation of poliovirus determines its neurotropism, although the molecular basis for this circumstance is not understood.
Compared with viral tissue tropism, the tissue tropism of bacterial and parasitic infections has not been as clearly elucidated, but studies of Neisseria species have provided insights. Both N. gonorrhoeae, which colonizes and infects the human genital tract, and N. meningitidis, which principally colonizes the human oropharynx but can spread to the brain, produce type IV pili (Tfp) that mediate adherence to host tissues. In the case of N. gonorrhoeae, the Tfp bind to a glucosamine-galactose-containing adhesin on the surface of cervical and urethral cells; in the case of N. meningitidis, the Tfp bind to cells in the human meninges and thus cross the blood-brain barrier. N. meningitidis expresses a capsular polysaccharide, while N. gonorrhoeae does not; however, there is no indication that this property plays a role in the different tissue tropisms displayed by these two bacterial species. N. gonorrhoeae can use cytidine monophosphate N-acetylneuraminic acid from host tissues to add N-acetylneuraminic acid (sialic acid) to its lipooligosaccharide O side chain, and this alteration appears to make the organism resistant to host defenses. Lactate, present at high levels on genital mucosal surfaces, stimulates sialylation of gonococcal lipooligosaccharide. Bacteria with sialic acid sugars in their capsules, such as N. meningitidis, E. coli K1, and group B streptococci, have a propensity to cause meningitis, but this generalization has many exceptions. For example, all recognized serotypes of group B streptococci contain sialic acid in their capsules, but only one serotype (III) is responsible for most cases of group B streptococcal meningitis. Moreover, both H. influenzae and S. pneumoniae can readily cause meningitis, but these organisms do not have sialic acid in their capsules.
TISSUE DAMAGE AND DISEASE
Disease is a complex phenomenon resulting from tissue invasion and destruction, toxin elaboration, and host response. Viruses cause much of their damage by exerting a cytopathic effect on host cells and inhibiting host defenses. The growth of bacterial, fungal, and protozoal parasites in tissue, which may or may not be accompanied by toxin elaboration, can compromise tissue function and lead to disease. For some bacterial and possibly some fungal pathogens, toxin production is one of the best-characterized molecular mechanisms of pathogenesis, while host factors such as IL-1, TNF-α, kinins, inflammatory proteins, products of complement activation, and mediators derived from arachidonic acid metabolites (leukotrienes) and cellular degranulation (histamines) readily contribute to the severity of disease.
Viral Disease Viral pathogens are well known to inhibit host immune responses by a variety of mechanisms. Immune responses can be affected by decreasing production of most major histocompatibility complex molecules (adenovirus E3 protein), by diminishing cytotoxic T cell recognition of virus-infected cells (Epstein-Barr virus EBNA1 antigen and cytomegalovirus IE protein), by producing virus-encoded complement receptor proteins that protect infected cells from complement-mediated lysis (herpesvirus and vaccinia virus), by making proteins that interfere with the action of IFN (influenza virus and poxvirus), and by elaborating superantigen-like proteins (mouse mammary tumor virus and related retroviruses and the rabies nucleocapsid). Superantigens activate large populations of T cells that express particular subsets of the T cell receptor β protein, causing massive cytokine release and subsequent host reactions. Another molecular mechanism of viral virulence involves the production of peptide growth factors for host cells, which disrupt normal cellular growth, proliferation, and differentiation. In addition, viral factors can bind to and interfere with the function of host receptors for signaling molecules. Modulation of cytokine production during viral infection can stimulate viral growth inside cells with receptors for the cytokine, and virus-encoded cytokine homologues (e.g., the Epstein-Barr virus BCRF1 protein, which is highly homologous to the immunoinhibitory IL-10 molecule) can potentially prevent immune-mediated clearance of viral particles. Viruses can cause disease in neural cells by interfering with levels of neurotransmitters without necessarily destroying the cells, or they may induce either programmed cell death (apoptosis) to destroy tissues or inhibitors of apoptosis to allow prolonged viral infection of cells. For infection to spread, many viruses must be released from cells. In a newly identified function, viral protein U (Vpu) of HIV facilitates the release of virus, a process that is specific to certain cells. Mammalian cells produce a restriction factor involved in inhibiting the release of virus; for HIV, this factor is designated BST-2 (bone marrow stromal antigen 2)/HM1.24/CD317, or tetherin. Vpu of HIV interacts with tetherin, promoting release of infectious virus. Overall, disruption of normal cellular and tissue function due to viral infection, replication, and release promotes clinical disease.
Bacterial Toxins Among the first infectious diseases to be understood were those due to toxin-elaborating bacteria. Diphtheria, botulism, and tetanus toxins are responsible for the diseases associated with local infections due to Corynebacterium diphtheriae, Clostridium botulinum, and Clostridium tetani, respectively. Clostridium difficile is an anaerobic gram-positive organism that elaborates two toxins, A and B, responsible for disruption of the intestinal mucosa when organism numbers expand in the intestine, leading to antibiotic-associated diarrhea and potentially to pseudomembranous colitis. Enterotoxins produced by E. coli, Salmonella, Shigella, Staphylococcus, and V. cholerae contribute to diarrheal disease caused by these organisms. Staphylococci, streptococci, P. aeruginosa, and Bordetella elaborate various toxins that cause or contribute to disease, including toxic shock syndrome toxin 1; erythrogenic toxin; exotoxins A, S, T, and U; and pertussis toxin. A number of bacterial toxins (e.g., cholera toxin, diphtheria toxin, pertussis toxin, E. coli heat-labile toxin, and P. aeruginosa exotoxin) have adenosine diphosphate ribosyl transferase activity; i.e., the toxins enzymatically catalyze the transfer of the adenosine diphosphate ribosyl portion of nicotinamide adenine diphosphate to target proteins and inactivate them. The staphylococcal enterotoxins, toxic shock syndrome toxin 1, and the streptococcal pyogenic exotoxins behave as superantigens, stimulating certain T cells to proliferate without processing of the protein toxin by antigen-presenting cells. Part of this process involves stimulation of the antigen-presenting cells to produce IL-1 and TNF-α, which have been implicated in many clinical features of diseases like toxic shock syndrome and scarlet fever. A number of gram-negative pathogens (Salmonella, Yersinia, and P. aeruginosa) can inject toxins directly into host target cells by means of a complex set of proteins referred to as the type III secretion system. Loss or inactivation of this virulence system usually greatly reduces the capacity of a bacterial pathogen to cause disease.
Endotoxin The lipid A portion of gram-negative LPS has potent biologic activities that cause many of the clinical manifestations of gram-negative bacterial sepsis, including fever, muscle proteolysis, uncontrolled intravascular coagulation, and shock. The effects of lipid A appear to be mediated by the production of potent cytokines due to LPS binding to CD14 and signal transduction via TLRs, particularly TLR4. Cytokines exhibit potent hypothermic activity through effects on the hypothalamus; they also increase vascular permeability, alter the activity of endothelial cells, and induce endothelial-cell procoagulant activity. Numerous therapeutic strategies aimed at neutralizing the effects of endotoxin are under investigation, but so far the results have been disappointing. It has been suggested that this lack of success may be due to substantial differences between mouse and human inflammatory responses to factors such as endotoxin; thus drugs developed in mouse models of infection may not be applicable to the human response.
Invasion Many diseases are caused primarily by pathogens growing in tissue sites that are normally sterile. Pneumococcal pneumonia is mostly attributable to the growth of S. pneumoniae in the lung and the attendant host inflammatory response, although specific factors that enhance this process (e.g., pneumolysin) may be responsible for some of the pathogenic potential of the pneumococcus. Disease that follows bloodstream infection and invasion of the meninges by meningitis-producing bacteria such as N. meningitidis, H. influenzae, E. coli K1, and group B streptococci appears to be due solely to the ability of these organisms to gain access to these tissues, multiply in them, and provoke cytokine production leading to tissue-damaging host inflammation.
Specific molecular mechanisms accounting for tissue invasion by fungal and protozoal pathogens are less well described. Except for studies pointing to factors like capsule and melanin production by C. neoformans and possibly levels of cell wall glucans in some pathogenic fungi, the molecular basis for fungal invasiveness is not well defined. Melanism has been shown to protect the fungal cell against death caused by phagocyte factors such as nitric oxide, superoxide, and hypochlorite. Morphogenic variation and production of proteases (e.g., the Candida aspartyl proteinase) have been implicated in fungal invasion of host tissues.
If pathogens are to effectively invade host tissues (particularly the blood), they must avoid the major host defenses represented by complement and phagocytic cells. Bacteria most often elude these defenses through their surface polysaccharides—either capsular polysaccharides or long O-side-chain antigens characteristic of the smooth LPS of gram-negative bacteria. These molecules can prevent the activation and/or deposition of complement opsonins or can limit the access of phagocytic cells with receptors for complement opsonins to these molecules when they are deposited on the bacterial surface below the capsular layer. Another potential mechanism of microbial virulence is the ability of some organisms to present the capsule as an apparent self antigen through molecular mimicry. For example, the polysialic acid capsule of group B N. meningitidis is chemically identical to an oligosaccharide found on human brain cells.
Immunochemical studies of capsular polysaccharides have led to an appreciation of the tremendous chemical diversity that can result from the linking of a few monosaccharides. For example, three hexoses can link up in more than 300 different, potentially serologically distinct ways, while three amino acids have only six possible peptide combinations. Capsular polysaccharides have been used as effective vaccines against meningococcal meningitis as well as against pneumococcal and H. influenzae infections and may prove to be of value as vaccines against any organisms that express a nontoxic, immunogenic capsular polysaccharide. In addition, most encapsulated pathogens become virtually avirulent when capsule production is interrupted by genetic manipulation; this observation emphasizes the importance of this structure in pathogenesis. It is noteworthy that the capsule-like surface polysaccharide PNAG has been found as a conserved structure shared by many microbes but generally is a poor target for antibody-mediated immunity because of the propensity of most humans and animals—all colonized by PNAG-producing microbes—to produce a nonprotective type of antibody. Altering the structure of PNAG by removing the acetate substituents on the N-acetylglucosamine monomers yields an immunogenic form, deacetylated PNAG, that reportedly induces antibodies that protect animals against diverse microbial pathogens.
Host Response The inflammatory response of the host is critical for interruption and resolution of the infectious process but is often responsible for the signs and symptoms of disease. Infection promotes a complex series of host responses involving the complement, kinin, and coagulation pathways. The production of cytokines such as IL-1, IL-18, TNF-α, IFN-γ, and other factors regulated in part by the NF-κB transcription factor leads to fever, muscle proteolysis, and other effects. An inability to kill or contain the microbe usually results in further damage due to the progression of inflammation and infection. For example, in many chronic infections, degranulation of host inflammatory cells can lead to release of host proteases, elastases, histamines, and other toxic substances that can degrade host tissues. Chronic inflammation in any tissue can lead to the destruction of that tissue and to clinical disease associated with loss of organ function, such as sterility from pelvic inflammatory disease caused by chronic infection with N. gonorrhoeae.
The nature of the host response elicited by the pathogen often determines the pathology of a particular infection. Local inflammation produces local tissue damage, while systemic inflammation, such as that seen during sepsis, can result in the signs and symptoms of septic shock. The severity of septic shock is associated with the degree of production of host effectors. Disease due to intracellular parasitism results from the formation of granulomas, wherein the host attempts to wall off the parasite inside a fibrotic lesion surrounded by fused epithelial cells that make up so-called multinucleated giant cells. A number of pathogens, particularly anaerobic bacteria, staphylococci, and streptococci, provoke the formation of an abscess, probably because of the presence of zwitterionic surface polysaccharides such as the capsular polysaccharide of Bacteroides fragilis. The outcome of an infection depends on the balance between an effective host response that eliminates a pathogen and an excessive inflammatory response that is associated with an inability to eliminate a pathogen and with the resultant tissue damage that leads to disease.
TRANSMISSION TO NEW HOSTS
As part of the pathogenic process, most microbes are shed from the host, often in a form infectious for susceptible individuals. However, the rate of transmissibility may not necessarily be high, even if the disease is severe in the infected individual, as these traits are not linked. Most pathogens exit via the same route by which they entered: respiratory pathogens by aerosols from sneezing or coughing or through salivary spread, gastrointestinal pathogens by fecal-oral spread, sexually transmitted diseases by venereal spread, and vector-borne organisms by either direct contact with the vector through a blood meal or indirect contact with organisms shed into environmental sources such as water. Microbial factors that specifically promote transmission are not well characterized. Respiratory shedding is facilitated by overproduction of mucous secretions, with consequently enhanced sneezing and coughing. Diarrheal toxins such as cholera toxin, E. coli heat-labile toxins, and Shigella toxins probably facilitate fecal-oral spread of microbial cells in the high volumes of diarrheal fluid produced during infection. The ability to produce phenotypic variants that resist hostile environmental factors (e.g., the highly resistant cysts of E. histolytica shed in feces) represents another mechanism of pathogenesis relevant to transmission. Blood parasites such as Plasmodium species change phenotype after ingestion by a mosquito—a prerequisite for the continued transmission of this pathogen. Venereally transmitted pathogens may undergo phenotypic variation due to the production of specific factors to facilitate transmission, but shedding of these pathogens into the environment does not result in the formation of infectious foci.
SUMMARY
In summary, the molecular mechanisms used by pathogens to colonize, invade, infect, and disrupt the host are numerous and diverse. Each phase of the infectious process involves a variety of microbial and host factors interacting in a manner that can result in disease. Recognition of the coordinated genetic regulation of virulence factor elaboration when organisms move from their natural environment into the mammalian host emphasizes the complex nature of the host-parasite interaction. Fortunately, the need for diverse factors in successful infection and disease implies that a variety of therapeutic strategies may be developed to interrupt this process and thereby to prevent and treat microbial infections.
146 |
Genomics and Infectious Disease |
Just as microscopy opened up the worlds of microbiology by providing a tool with which to visualize microorganisms, technological advances in genomics are now providing microbiologists with powerful new methods with which to characterize the genetic map underlying all microbes with unprecedented resolution, thereby illuminating their complex and dynamic interactions with one another, the environment, and human health. The field of infectious disease genomics encompasses a vast frontier of active research that has the potential to transform clinical practice in relation to infectious diseases. While genetics has long played a key role in elucidating the process of infection and managing clinical infectious diseases, the ability to extend our thinking and our approaches beyond the study of single genes to an examination of the sequence, structure, and function of entire genomes is identifying new possibilities for research and opportunities to change clinical practice. From the development of diagnostics with unprecedented sensitivity, specificity, and speed to the design of novel public health interventions, technical and statistical genomic innovations are reshaping our understanding of the influence of the microbial world on human health and providing us with new tools to combat infection. This chapter explores the application of genomics methods to microbial pathogens and the infections they cause (Table 146-1). It discusses innovations that are driving the development of diagnostic approaches and the discovery of new pathogens; providing insight into novel therapeutic approaches and paradigms; and advancing methods in infectious disease epidemiology and the study of pathogen evolution that can inform infection control measures, public health responses to outbreaks, and vaccine development. We draw on examples in current practice and from the recent scientific literature as signposts that point toward the ways in which the insights from pathogen genomics may influence infectious diseases in the short and long terms. Table 146-2 provides definitions for a selection of important terms used in genomics.
|
CURRENT CLINICAL APPLICATIONS OF INFECTIOUS DISEASE GENOMICS |
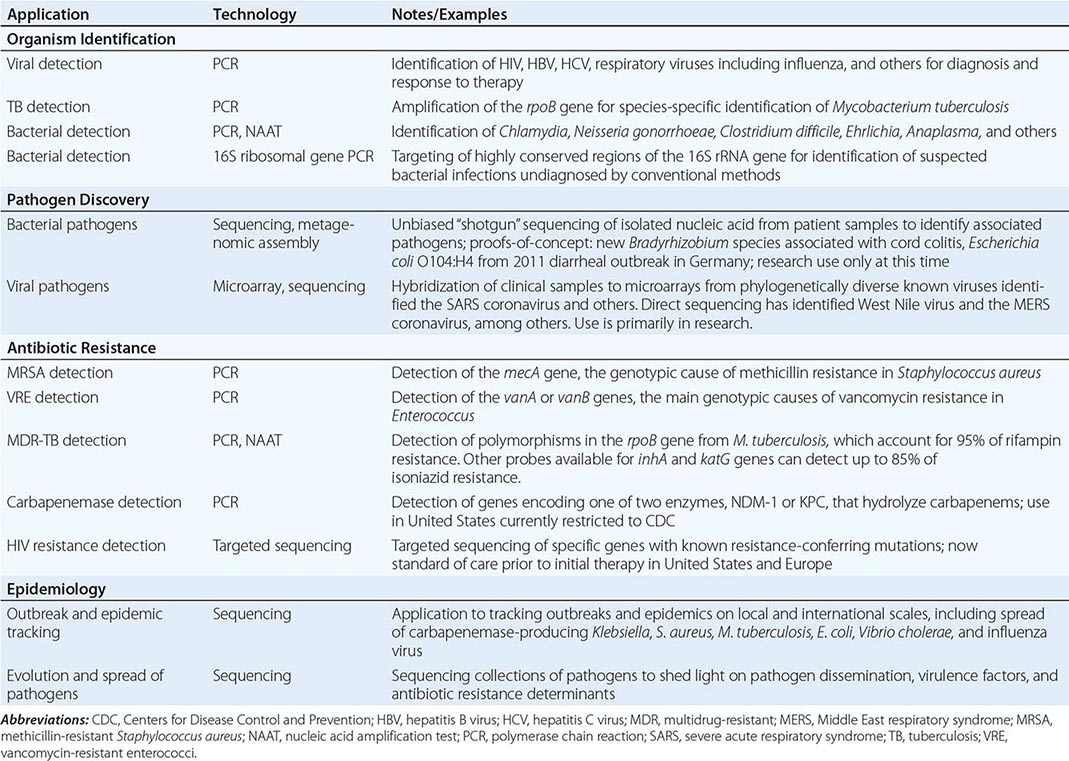
|
GLOSSARY OF SELECTED TERMS IN GENOMICS |
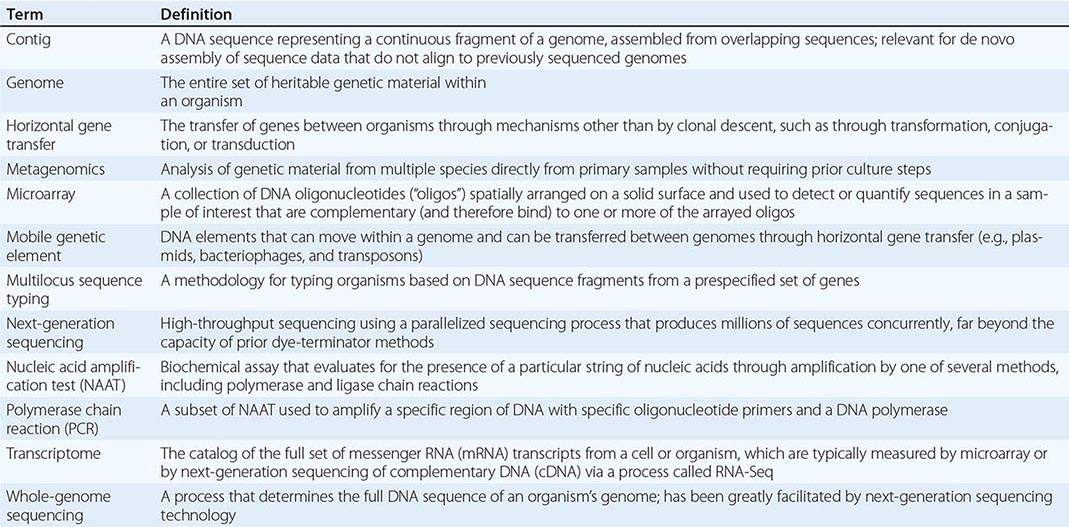
MICROBIAL DIAGNOSTICS
The basic goals of a clinical microbiology laboratory are to establish the presence of a pathogen in a clinical sample, to identify the pathogen, and, when possible, to provide other information that can help guide clinical management and even prognosis, such as antibiotic susceptibility profiles or the presence of virulence factors. To date, clinical microbiology laboratories have largely approached these goals phenotypically by growth-based assays and biochemical testing. Bacteria, for instance, are algorithmically grouped into species by their characteristic microscopic appearance, nutrient requirements for growth, and ability to catalyze certain reactions. Antibiotic susceptibility is determined in most cases by assessing growth in the presence of antibiotic.
With the sequencing revolution paving the way to easy access of complete pathogen genomes (Fig. 146-1), we are now able to more systematically clarify the genetic basis of these observable phenotypes. Compared with traditional growth-based methods for bacterial diagnostics that dominate the clinical microbiology laboratory, nucleic acid–based diagnostics promise improved speed, sensitivity, specificity, and breadth of information. Bridging clinical and research laboratories, adaptations of genomic technologies have begun to deliver on this promise.
FIGURE 146-1 Completed bacterial genome sequences by year, through 2012. (Data compiled from www.genomesonline.org.)
HISTORICAL LIMITATIONS AND PROGRESS THROUGH GENETIC APPROACHES
The molecular diagnostics revolution in the clinical microbiology laboratory is well under way, borne of necessity in the effort to identify microbes that are refractory to traditional culture methods. Historically, diagnosis of many so-called unculturable pathogens has relied largely on serology and antigen detection. However, these methods provide only limited clinical information because of their suboptimal sensitivity and specificity as well as the long delays that diminish their utility for real-time patient management. Newer tests to detect pathogens based on nucleic acid content have already offered improvements in the select cases to which they have been applied thus far.
Unlike direct pathogen detection, serologic diagnosis—measurement of the host’s response to pathogen exposure—can typically be made only in retrospect, requiring both acute- and convalescent-phase sera. For chronic infections, distinguishing active from latent infection or identifying repeat exposure by serology alone can be difficult or impossible, depending on the syndrome. In addition, the sensitivity of serologic diagnosis varies with the organism and the patient’s immune status. For instance, tuberculosis is notoriously difficult to identify by serologic methods; tuberculin skin testing using purified protein derivative (PPD) is especially insensitive in active disease and may be cross-reactive with vaccines or other mycobacteria. Even the newer interferon γ release assays (IGRAs), which measure cytokine release from T lymphocytes in response to Mycobacterium tuberculosis–specific antigens in vitro, have limited sensitivity in immunodeficient hosts. Neither PPD testing nor IGRAs can distinguish latent from active infection. Serologic Lyme disease diagnostics suffer similar limitations: in patients from endemic regions, the presence of IgG antibodies to Borrelia burgdorferi may reflect prior exposure rather than active disease, while IgM antibodies are imperfectly sensitive and specific (50% and 80%, respectively, in early disease). The complex nature of these tests, particularly in view of the nonspecific symptoms that may accompany Lyme disease, has had substantial implications on public perceptions of Lyme disease and antibiotic misuse in endemic areas. Similarly, syphilis, a chronic infection caused by Treponema pallidum, is notoriously difficult to stage by serology alone, requiring the use of multiple different nontreponemal (e.g., rapid protein reagin) and treponemal (e.g., fluorescent treponemal antibody) tests in conjunction with clinical suspicion. Complementing serology, antigen detection can improve sensitivity and specificity in select cases but has been validated only for a limited set of infections. Typically, structural elements of pathogens are detected, including components of viral envelopes (e.g., hepatitis B surface antigen, HIV p24 antigen), cell surface markers in certain bacteria (e.g., Streptococcus pneumoniae, Legionella pneumophila serotype 1) or fungi (e.g., Cryptococcus, Histoplasma), and less specific fungal cell-wall components such as galactomannan and β-glucan (e.g., Aspergillus and other dimorphic fungi).
Given the impracticality of culture and the lack of sensitivity or sufficient clinical information afforded by serologic and antigenic methods, the push toward nucleic acid–based diagnostics originated in pursuit of viruses and fastidious bacteria, becoming part of the standard of care for select organisms in U.S. hospitals. Such tests, including polymerase chain reaction (PCR) and other nucleic acid amplification tests (NAATs), are now widely used for many viral infections, both chronic (e.g., HIV infection) and acute (e.g., influenza). This technique provides essential information about both the initial diagnosis and the response to therapy and in some cases genotypically predicts drug resistance. Indeed, progression from antigen detection to PCR transformed our understanding of the natural course of HIV infection, with profound implications for treatment (Fig. 146-2). In the early years of the AIDS pandemic, p24 antigenemia was detected in acute HIV infection but then disappeared for years before emerging again with progression to AIDS (Fig. 146-2B). Without a marker demonstrating viremia, the role of treatment during HIV infection prior to the development of clinical AIDS was uncertain, and monitoring treatment efficacy was challenging. With the emergence of PCR as a progressively more sensitive test (now able to detect as few as 20 copies of virus per milliliter of blood), viremia was recognized as a near-universal feature of HIV infection. This recognition has been transformative in guiding the initiation of therapy as well as adjustments in therapy and, together with the development of less toxic therapies, has helped to shape guidelines that now favor earlier introduction of antiretroviral therapy for HIV infection.
FIGURE 146-2 A. Timeline of select milestones in HIV management. Genomic advances are shown in bold type. The approvals and recommendations indicated apply to the United States. ARV, antiretroviral; AZT, zidovudine; NRTI, nucleoside reverse transcriptase (RT) inhibitor; NNRTI, non-nucleoside RT inhibitor; PI, protease inhibitor. B. Viral dynamics in the natural history of HIV infection. Three diagnostic markers are shown: HIV antibody (Ab), p24 antigen (p24), and viral load (VL). Dashed gray line represents limit of detection. (Adapted from data in HH Fiebig et al: Dynamics of HIV viremia and antibody seroconversion in plasma donors: Implications for diagnosis and staging of primary HIV infection. AIDS 17:1871, 2003.)
As they are for viruses, nucleic acid–based tests have become the diagnostic tests of choice for fastidious bacteria, including the common sexually transmitted intracellular bacterial pathogens Neisseria gonorrhoeae and Chlamydia trachomatis as well as the tick-borne Ehrlichia chaffeensis and Anaplasma phagocytophilum. More recently, nucleic acid amplification–based detection has offered improved sensitivity for diagnosis of the important nosocomial pathogen Clostridium difficile; NAATs can provide clinically relevant information on the presence of cytotoxins A and B as well as molecular markers of hypervirulence such as those characterizing the recently recognized North American pulsotype 1 (NAP1), which is found more frequently in cases of severe illness. The importance of genomics in selecting loci for diagnostic assays and in monitoring test sensitivity was recently highlighted by the emergence in Sweden of a new variant of C. trachomatis containing a deletion that includes the gene targeted by a set of commercial NAATs. By evading detection through this deletion (which would have prompted the initiation of treatment), this strain came to be highly prevalent in some areas of Sweden. While nucleic acid–based tests remain the diagnostic approach of choice for fastidious bacteria, this example serves as a reminder of the need for careful development and ongoing monitoring of molecular diagnostics.
In contrast, for typical bacterial pathogens for which culture methods are well established, growth-based assays followed by biochemical tests still dominate in the clinical laboratory. Informed by decades of clinical microbiology, these tests have served clinicians well, yet the limitations of growth-based tests—in particular, the delays associated with waiting for growth—have left open opportunities for improvements. Molecular diagnostics, greatly informed by the vast quantity of microbial genome sequences generated in recent years, offers a way forward. First, sequencing studies may identify key genes (or noncoding nucleic acids) that can be developed into targets for clinical assays using PCR or hybridization platforms. Second, sequencing itself may eventually become inexpensive and rapid enough to be performed routinely on clinical specimens, with consequent unbiased detection of pathogens.
ORGANISM IDENTIFICATION
In order to adapt nucleic acid detection to diagnostic tests and thus to identify pathogens on a wide scale, sequences must be identified that are conserved enough within a species to identify the diversity of strains that may be encountered in various clinical settings, yet divergent enough to distinguish one species from another. Until recently, this problem has been solved for bacteria by targeting the element of a bacterial genome that is most highly conserved within a species: the 16S ribosomal RNA (rRNA) subunit. At present, 16S PCR amplification from tissue specimens can be performed by specialty laboratories, though its sensitivity and clinical utility to date have remained somewhat limited because, for instance, of inhibitory molecules often found in clinical tissue samples that prevent reliable, sensitive PCR amplification. As such barriers are reduced through technological advances and as the causes of culture-negative infection are clarified (perhaps in part through sequencing efforts), these tests may become both more accessible and more helpful.
With the wealth of sequencing data now available, other regions beyond 16S rRNA can be targeted for bacterial species identification. These other genomic loci can provide additional information about a clinical isolate that is relevant to patient management. For instance, detection of the presence—or potentially even the expression—of toxin genes such as those for C. difficile toxins A and B or Shiga toxin may provide clinicians with additional information that will help distinguish commensals or colonizing bacteria from pathogens and thus aid in prognostication as well as diagnosis.
While amplification tests such as PCR exemplify one approach to nucleic acid detection, other approaches exist, including detection by hybridization. Although not currently used in the clinical realm, techniques for detection and identification of pathogens by hybridization to microarrays are being developed for other purposes. Of note, these different detection techniques require different degrees of conservation. Highly sensitive amplification methods require a high degree of sequence identity between PCR primer pairs and their short, specific target sequences; even a single base-pair mismatch (particularly near the 3′ end of the primer) may interfere with detection. In contrast, hybridization-based tests are more tolerant of mismatch and thus can be used to detect important regions that may be less precisely conserved within a species, thus potentially allowing detection of clinical isolates from a given species with greater diversity between isolates. Such assays take advantage of the predictable binding interactions of nucleic acids. The applicability of hybridization-based methods toward either DNA or RNA opens up the possibility of expression profiling, which can uncover phenotypic information from nucleic acid content.
Both PCR and hybridization methods target specific, known organisms. At the other extreme, as sequencing costs and turnaround times decrease, direct metagenomic sequencing from patient samples is becoming increasingly feasible. This shotgun sequencing approach is unbiased—i.e., is able to detect any microbial sequence, however divergent or unexpected. This new approach brings its own set of challenges, however, including the need to recognize pathogenic sequences against a background of expected host and commensal sequences and to distinguish true pathogens from either colonizers or laboratory contaminants. In a powerful example of this new frontier of sequencing-based clinical diagnosis, investigators diagnosed neuroleptospirosis in a child with an unexplained encephalitis syndrome by finding sequences corresponding to the Leptospira genus in cerebrospinal fluid from the patient. Rapid (<48-h) sequencing and analysis informed the patient’s care in real time, leading to life-saving targeted antibiotic therapy for an unexpected diagnosis that was impossible to make through standard laboratory testing. The diagnosis was retrospectively confirmed through both convalescent serologies and PCR using primers designed on the basis of sequencing data.
PATHOGEN DISCOVERY
In addition to clinical diagnostic applications, novel genomic technologies, including whole-genome sequencing, are being applied to clinical research specimens with a goal of identifying new pathogens in a variety of circumstances. The tremendous sensitivity and unbiased nature of sequencing is also ideal in searching clinical samples for unknown or unsuspected pathogens.
Causal inference in infectious diseases has progressed since the time of Koch, whose historical postulates provided a rigorous framework for attributing a disease to a microorganism. According to an updated version of Koch’s postulates, an organism, whether it can be cultured or not, should induce disease upon introduction into a healthy host if it is to be implicated as a causative pathogen. Current sequencing technologies are ideal for advancing this modern version of Koch’s postulates because they can identify candidate causal pathogens with unprecedented sensitivity and in an unbiased way, unencumbered by limitations such as culturability. Yet, as direct sequencing on primary patient samples greatly expands our ability to recognize associations between microbes and disease states, critical thinking and experimentation will continue to be vital to establishing causality.
Virus discovery in particular has been greatly facilitated by new nucleic acid technology. These frontiers were first notably explored with high-density microarrays containing spatially arrayed sequences from a phylogenetically diverse collection of viruses. Although biased toward those with homology to known viruses, novel viruses in clinical samples were successfully identified on the basis of their ability to hybridize to these prespecified sequences. This methodology famously contributed to identification of the coronavirus causing severe acute respiratory syndrome (SARS). Once discovered, this SARS coronavirus was rapidly sequenced: the full genome was assembled in April 2003, less than 6 months after recognition of the first case. This accomplishment illustrated the advancing power and speed of new diagnostic technologies.
With the advent of next-generation sequencing, unbiased pathogen discovery is now possible through a process known as metagenomic assembly (Fig. 146-3). Sequences of random nucleotide fragments can be generated from clinical specimens with no a priori knowledge of pathogen identity through a process called shotgun sequencing. This collection of sequences can then be computationally aligned to host (i.e., human) sequences, with aligned sequences removed and remaining sequences compared with other known genomes to detect the presence of known microorganisms. Sequence fragments that remain unaligned suggest the presence of an additional organism that cannot be matched to a known, characterized genome; these reads can be assembled into contiguous nucleic acid stretches that can be compared to known sequences to construct the genome of a potentially novel organism. Assembled genomes (or parts of genomes) can then be compared to known genomes to infer the phylogeny of new organisms and identify related classes or traits. Thus, not only can this process identify unanticipated pathogens; it can even identify undiscovered organisms. Some early applications of sequencing on clinical samples have centered around the discovery of novel viruses, including such emerging pathogens as West Nile virus, SARS coronavirus, and the Middle East respiratory syndrome coronavirus (MERS-CoV) that has caused severe respiratory illnesses in healthy adults, as well as viral causes of myriad other conditions, from tropical hemorrhagic fevers to diarrhea in newborns.
FIGURE 146-3 Workflow of metagenomic assembly for pathogen discovery. DNA is isolated from a specimen of interest (e.g., tissue, body fluid) containing a mixture of host DNA and nucleic acids from coexisting microbes, either commensal or pathogenic. All DNA (and RNA if a reverse transcription step is added) is then sequenced, yielding a mixture of DNA sequence fragments (“reads”) from organisms present. These reads are then aligned to existing reference genomes for the host or any known microbes, leaving reads that do not align (“map”) to any known sequence. These unmapped reads are then computationally assembled de novo into the largest contiguous stretches of DNA possible (“contigs”), representing fragments of previously unsequenced genomes. These genome fragments (contigs) are then mapped onto a phylogenetic tree based on their sequence. Some may represent known but as-yet-unsequenced organisms, while others will represent novel species. (Figure prepared with valuable input from Dr. Ami S. Bhatt, personal communication.)
More recently, metagenomic assembly has been successfully extended to bacterial pathogen discovery. Investigators identified a new bacterial species associated with “cord colitis”—a rare antibiotic-responsive, culture-negative colitis in recipients of umbilical cord-blood stem cells—by sequencing colon biopsy samples from affected patients and matched controls. A single dominant species emerged from metagenomic assembly in samples from patients that was absent from control samples. The presence of this species was confirmed by PCR and fluorescence in situ hybridization on primary tissue samples. On the basis of its similarity to other known species, the organism was named Bradyrhizobium enterica, a novel species from a genus that has proved difficult to culture and thus would have been hard to identify by other means. Correlation versus causation remains an open question; therefore, further efforts will be required to make such links.
As metagenomic sequencing and assembly techniques become more robust, this technology holds great promise for identifying microorganisms that are associated with clinical conditions of unknown etiology. Conventional methods already have unexpectedly linked numerous conditions with specific agents of infection—e.g., cervical and oropharyngeal cancers with human papillomavirus, Kaposi’s sarcoma with human herpesvirus 8, and certain lymphomas with Epstein-Barr virus. Sequencing techniques offer unprecedented sensitivity and specificity for identifying foreign nucleic acid sequences that may suggest other conditions—from malignancies to inflammatory conditions to unexplained fevers or other clinical syndromes—associated with organisms from viruses to bacteria to parasites. As sequencing-based discovery expands, microbes may be found to be associated with conditions not classically thought of as infectious. Studies of bowel flora in laboratory animals and even humans are already beginning to suggest correlations between microbe composition and various aspects of metabolic and cardiovascular health. Improved methods for pathogen detection will continue to uncover unexpected correlations between microbes and disease states, but the mere presence of a microbe does not establish causality. Fortunately, once the relatively laborious and computationally intensive metagenomic sequencing and assembly efforts have identified a pathogen, further detection can easily be undertaken with targeted methods such as PCR or hybridization, which are much more straightforward and scalable. This capacity should facilitate the additional careful investigation that will be required to progress beyond correlation and to draw causal inference.
ANTIBIOTIC RESISTANCE
At present, antibiotic resistance in bacteria and fungi is determined by isolating a single colony from a cultured clinical specimen and testing its growth in the presence of a drug. The requirement for multiple growth steps in these conventional assays has several consequences. First, only culturable pathogens can be readily processed. Second, this process requires considerable infrastructure to support the sterile environment required for culture-based testing of diverse organisms. Finally, and perhaps most significantly, even the fastest-growing organisms require 1–2 days of processing for identification and 2–3 days for determination of susceptibilities. Slower-growing organisms take even longer: for instance, weeks must pass before drug-resistant M. tuberculosis can be identified by growth phenotype. Given the clinical imperative in serious illness to begin effective therapy early, this inherent delay in susceptibility determination has obvious implications for empirical antibiotic use: broad-spectrum antibiotics often must be chosen up front in situations where it is later shown that preferred narrower-spectrum drugs would have been effective or even that no antibiotics were appropriate (i.e., in viral infections). With this strategy, the empirical choice can be incorrect, often with devastating consequences. Real-time identification of the infecting organism and information on its susceptibility profile would guide initial therapy and support judicious antibiotic use, ideally improving patient outcomes while aiding in the ever-escalating struggle with antibiotic resistance by reserving the use of broad-spectrum agents for cases in which they are truly needed.
Molecular diagnostics and sequencing offer a way to accelerate detection of a pathogen’s antibiotic susceptibility profile. If a genotype that confers resistance can be identified, this genotype can be targeted for molecular detection. In infectious disease, this approach has most convincingly come to fruition for HIV (Fig. 146-2A). (In a conceptually parallel application of genomic analysis, molecular detection of certain resistance determinants in cancers is beginning to inform chemotherapeutic selection.) Extensive sequencing of HIV strains and correlations drawn between viral genotypes and phenotypic resistance have delineated the majority of mutations in key HIV genes, such as reverse transcriptase, protease, and integrase, that confer resistance to the antiretroviral agents that target these proteins. For instance, the single-amino-acid substitution K103N in the HIV reverse transcriptase gene predicts resistance to the first-line nonnucleoside reverse transcriptase inhibitor efavirenz, and its detection thus informs a clinician to choose a different agent. The effects of these common mutations on HIV susceptibility to various drugs—as well as on viral fitness—are curated in publically available databases. Thus, genotypes are now routinely used to predict drug resistance in HIV, as phenotypic resistance assays are far more cumbersome than targeted sequencing. Indeed, current recommendations in the United States are to sequence virus from a patient’s blood before initiating antiretroviral therapy, which is then tailored to the predicted resistance phenotype. As new targeted therapies are introduced, this targeted sequencing–based approach to drug resistance will likely prove important in other viral infections (e.g., hepatitis C).
For several reasons, the challenge of predicting antibiotic susceptibility from genotype has not yet been met in bacteria to the same degree as in HIV. In general, bacteria have evolved diverse resistance mechanisms to most antibiotics; thus, the task cannot be reduced to probing for a single genetic lesion, target, or mechanism. For instance, at least five distinct modes of resistance to fluoroquinolones are known: reduced import, increased efflux, mutated target sites, drug modification, and shielding of the target sites by expression of another protein. Further, we lack a comprehensive compendium of genetic elements conferring resistance, and new mechanisms and genes emerge regularly in the face of antibiotic deployment. As bacteria have far more complex genomes than viruses, with thousands of genes on their chromosomes and the capacity for acquiring many more through horizontal gene transfer of plasmids and mobile genetic elements within and even between species, the task of not only defining all current but also predicting all future mechanisms at a genetic level is daunting or perhaps impossible.
Despite these challenges, in a few select cases where the genotypic basis for resistance has been well defined, genotypic assays for antibiotic resistance are already being introduced into clinical practice. One important example is the detection of methicillin-resistant Staphylococcus aureus (MRSA). S. aureus is one of the most common and serious bacterial pathogens of humans, particularly in health care settings. Resistance to methicillin, the most effective antistaphylococcal antibiotic, has become very common even in community-acquired strains. The alternative to methicillin, vancomycin, is effective against MRSA but measurably inferior to methicillin against methicillin-susceptible S. aureus (MSSA). Analysis of clinical MRSA isolates has demonstrated that the molecular basis for resistance to methicillin in essentially all cases stems from the expression of an alternative penicillin-binding protein (PBP2A) encoded by the gene mecA, which is found within a transferable genetic element called mec. This mobile cassette has spread rapidly through the S. aureus population via horizontal gene transfer and selection from widespread antibiotic use. Because resistance is essentially always due to the presence of the mec cassette, MRSA is amenable to molecular detection. In recent years, a PCR test for the presence of the mec cassette, which saves hours to days compared with standard culture-based methods, has been approved by the U.S. Food and Drug Administration.
Additional molecular diagnostics are being implemented in the evaluation of bacterial antibiotic resistance. Vancomycin-resistant enterococci (VRE) harbor one of a limited number of van genes responsible for resistance to this important antibiotic by altering the mechanism for cell wall cross-linking that vancomycin inhibits. Detection of one of these genes by PCR indicates resistance. Identification of two carbapenemase-encoding plasmids—NDM-1 and KPC—responsible for a significant fraction of carbapenem resistance (though not for all such resistance) has led to the development of a multiplexed PCR assay to detect these important resistance elements. Because carbapenems are broad-spectrum antibiotics frequently reserved for multidrug-resistant bacteria (particularly enteric gram-negative bacilli) and are often used as antibiotics of last resort, the initial appearance of resistance and the subsequent increase in its prevalence have caused considerable concern. Therefore, even though other mechanisms for carbapenem resistance exist, a rapid PCR test for the two plasmids encoding these two carbapenemases has been developed to aid in both diagnostic and infection control efforts. Efforts are under way to extend this multiplexed PCR assay to other plasmid-borne carbapenemases and thus to make it more comprehensive.
The power and application of molecular genetic tests are not limited to high-income settings. With the increasing burden of drug-resistant tuberculosis in the developing world, a molecular diagnostic test has now been developed to detect rifampin-resistant tuberculosis. The genetic basis for rifampin resistance has been well defined by targeted sequencing: characteristic mutations in the molecular target of rifampin, RNA polymerase, account for the vast majority of rifampin-resistant strains of M. tuberculosis. A PCR assay that can detect both the M. tuberculosis organism and a rifampin-resistant allele of RNA polymerase from clinical samples has recently been approved. Since rifampin resistance frequently accompanies resistance to other antibiotics, this test can suggest the possible presence of multidrug-resistant M. tuberculosis within hours instead of weeks.
Despite differences in relative genome complexity, HIV genotypic screening for antiretroviral resistance offers one framework for broadening efforts at genotypic assays for nonviral antibiotic resistance. As whole-genome pathogen sequencing has become increasingly feasible and inexpensive (Fig. 146-1), significant efforts have been launched to sequence hundreds to thousands of antibiotic-sensitive and -resistant isolates of a given pathogen in order to more comprehensively define resistance-conferring genetic elements. In parallel with advancing sequencing technologies, progress in computational techniques, bioinformatics and statistics, and data storage as well as experimental confirmatory testing of hypotheses will be needed to move toward the ambitious goal of a comprehensive compendium of antibiotic resistance determinants. Open sharing and careful curation of new sequence information will be of paramount importance.
Yet no matter how thorough and carefully curated such a genotype-phenotype database is, history suggests that comprehensively cataloguing resistance in nonviral pathogens, with new mechanisms continuously emerging, will be challenging at best. Even identifying and itemizing current resistance mutations is a daunting prospect: nonviral genomes are much larger than viral ones, and their abundance and diversity are such that hundreds to thousands of genetic differences often exist between clinical isolates, of which perhaps only one may cause resistance. For example, increasing resistance to artemisinin, one of the most effective new agents for malaria, has prompted recent large-scale efforts to identify the basis for resistance. While such studies have identified promising leads, no clear mechanism has emerged; in fact, a single genetic lesion alone may not fully account for resistance. Especially with multiple possible resistance mechanisms for a given antibiotic as well as ongoing evolutionary pressure resulting in the development and acquisition of new modes of resistance, a genotypic approach to diagnosing antibiotic resistance is likely to be imperfect.
We have already observed the accumulation of new or unanticipated modes of resistance from ongoing evolutionary pressure caused by the widespread clinical use of antibiotics. Even with MRSA, perhaps the best-studied case of antibiotic resistance and a model of relative simplicity with a single known monogenic resistance determinant (mecA), a genotype-based approach to resistance detection proved flawed. One limitation was a recall of the initial commercial genotypic resistance assay that was deployed for the identification of MRSA. A clinical isolate of S. aureus emerged in Belgium that expressed a variant of the mec cassette not detected by the assay’s PCR primers. New primers were added to detect this new variant, and the assay was re-approved for use. More recently, an even more divergent but functionally analogous gene called mecC, which confers methicillin resistance but evades PCR detection by this assay, was found. This series of events exemplifies the need for ongoing monitoring of any genotypic resistance assay. A second limitation is that a contradiction can occur between genotypic and phenotypic evidence for resistance. Up to 5% of MSSA strains carry a copy of the mecA gene that is either nonfunctional or not expressed. Thus, the erroneous identification of these strains as MRSA by genotypic detection would lead to administration of the inferior antibiotic vancomycin rather than the preferred β-lactam therapy.
These examples illustrate one of the prime challenges of moving beyond growth-based assays: genotype is merely a proxy for the resistance phenotype that directly informs patient care. One alternative approach currently under development attempts to circumvent the limitations of genotypic resistance testing by returning to a phenotypic approach, albeit one informed by genomic methods: transcriptional profiles serve as a rapid phenotypic signature for antibiotic response. Conceptually, since dying cells are transcriptionally distinct from cells fated to survive, susceptible bacteria enact different transcriptional profiles after antibiotic exposure that are different from the profiles of resistant strains, independent of the mechanism of resistance. These differences can be measured and, since transcription is one of the most rapid responses to cell stress (minutes to hours), can be used to determine whether cells are resistant or susceptible much more rapidly than is possible if one waits for growth in the presence of antibiotics (days). Like DNA, RNA can be readily detected through predictable rules governing base pairing via either amplification or hybridization-based methods. Changes in a carefully selected set of transcripts form an expression signature that can represent the total cellular response to antibiotic without requiring full characterization of the entire transcriptome. Preliminary proof-of-concept studies suggest that this approach may identify antibiotic susceptibility on the basis of transcriptional phenotype much more quickly than is possible with growth-based assays.
Because of its sensitivity in detecting even very rare nucleic acid fragments, sequencing is now permitting studies of unprecedented depth into complex populations of cells and tissues. The strength of this depth and sensitivity applies not only to the detection of rare, novel pathogens in a sea of host signal but also to the identification of heterogeneous pathogen subpopulations in a single host that may differ, for example, in drug resistance profiles or pathogenesis determinants. Future studies will be needed to elucidate the clinical significance of these variable subpopulations, even as deep sequencing is now providing unprecedented levels of detail about majority and minority members of this population.
HOST-BASED DIAGNOSTICS
While pathogen-based diagnostics continue to be the mainstay for diagnosing infection, serologic testing has long been the basis of a strategy to diagnose infection by measuring host responses. Here, too, the application of genomics is now being explored to improve upon this approach, given the previously described limitations of serologic testing. Rather than using antibody responses as a retrospective biomarker for infection, recent efforts have focused on transcriptomic analysis of the host response as a new direction with diagnostic implications for human disease. For instance, while pathogen-based diagnostic tests to distinguish active from latent tuberculosis infection have proved elusive, recent work shows that the transcriptional profile of circulating white blood cells exhibits a differential pattern of expression of nearly 400 transcripts that distinguish active from latent tuberculosis; this expression pattern is driven in part by changes in interferon-inducible genes in the myeloid lineage. In a validation cohort, this transcriptional signature was able to distinguish patients with active versus latent disease, to distinguish tuberculosis infection from other pulmonary inflammatory states or infections, and to track responses to treatment in as little as 2 weeks, with normalization of expression toward that of patients without active disease over 6 months of effective therapy. Such a test could play an important role not only in the management of patients but also as a marker of efficacy in clinical trials of new therapeutic agents. Similarly, other investigators have been trying to identify host transcriptional signatures in circulating blood cells that are distinct in influenza A infection from those in upper respiratory infections caused by certain other viruses or bacteria. These signatures also varied with phase of infection and showed promise in distinguishing exposed subjects who will become symptomatic from those who will not. These results suggest that profiling of host transcriptional dynamics could augment the information obtained from studies of pathogens, both enhancing diagnosis and monitoring the progression of illness and the response to therapy.
In this era of genome-wide association studies and attempts to move toward personalized medicine, genomic approaches are also being applied to the identification of host genetic loci and factors that contribute to infection susceptibility. Such loci will have undergone strong selection among populations in which the disease is endemic. By identifying the beneficial genetic alleles among individuals who survive in such settings, markers for susceptibility or resistance are being discovered; these markers can be translated into diagnostic tests to identify susceptible individuals in order to implement preventive or prophylactic interventions. Further, such studies may offer mechanistic insight into the pathogenesis of infection and inform new methods of therapeutic intervention. Such beneficial genetic associations were recognized long before the advent of genomics, as in the protective effects of the negative Duffy blood group or heterozygous hemoglobin abnormalities against Plasmodium infection. Genomic methods enable more systematic and widespread investigations of the host to identify not only people with altered susceptibility to numerous diseases (e.g., HIV infection, tuberculosis, and cholera) but also host factors that contribute to and thus might predict the severity of disease.
THERAPEUTICS
Genomics has the potential to impact infectious disease therapeutics in two ways. By transforming the speed of diagnostic information acquisition or the type of diagnostic information that can be attained, it can influence therapeutic decision-making. Alternatively, by opening up new avenues to understanding pathogenesis, providing new ways to disrupt infection, and delineating new approaches to antibiotic discovery, it can facilitate the development of new therapeutic agents.
GENOMIC DIAGNOSTICS INFORMING THERAPEUTICS
Efforts at antibiotic discovery are declining, with few new agents in the pipeline and even fewer entering the market. This phenomenon is due in part to the lack of economic incentives for the private sector; however, it is also attributable in part to the enormous challenges involved in the discovery and development of antibiotics. For obvious market-related reasons, nearly all efforts have focused on broad-spectrum antibiotics; the development of a chemical entity that works across an extremely diverse set of organisms (i.e., more divergent from each other than a human is from an amoeba) is far more challenging than the development of an agent that is designed to target a single bacterial species. Nevertheless, the concept of narrow-spectrum antibiotics has heretofore been rejected because of the lack of early diagnostic information that would guide the selection of such agents. Thus, rapid diagnostics providing antibiotic susceptibility information that can guide antibiotic selection in real time have the potential to alter and simplify antibiotic strategies by allowing a paradigm shift away from broad-spectrum drugs and toward narrow-spectrum agents. Such a paradigm shift clearly would have additional implications for the current escalation of antibiotic resistance.
In yet another diagnostic paradigm with the potential to impact therapeutic interventions, genomics is opening new avenues to a better understanding not only of different host susceptibilities to infection but also of different host responses to therapy. In a sense, the promise of “personalized medicine” has been a tantalizing holy grail. Some signs now point to the realization of this goal. For example, the role of glucocorticoids in tuberculous meningitis has been long debated. Recently, polymorphisms in the human genetic locus LTA4H, which encodes a leukotriene-modifying enzyme, were found to modulate the inflammatory response to tuberculosis. Patients with tuberculous meningitis who were homozygous for the proinflammatory LTA4H allele were most helped by adjunctive glucocorticoid treatment, while those who were homozygous for the anti-inflammatory allele were negatively affected by steroid treatment. This anti-inflammatory adjunct has become the standard of care in tuberculous meningitis, but this study suggests that perhaps only a subset of patients benefit and further suggests a genetic means of prospectively identifying this subset. Thus, genomic diagnostic tests may eventually inform diagnosis, prognosis, and treatment decisions by revealing the pathogenic potential of the microbe and detecting host responses to both infection and therapy.
GENOMICS IN DRUG AND VACCINE DEVELOPMENT
Genomic technologies are already dramatically changing research on host–pathogen interactions, with a goal of increasingly influencing the process of therapeutic discovery and development. Sequencing offers several possible avenues into antimicrobial therapeutic discovery. First, genomic-scale molecular methods have paved the way for comprehensive identification of all essential genes encoded within a pathogen’s genome, with consequent systematic identification of all possible vulnerabilities within a pathogen that could be targeted therapeutically. Second, transcriptional profiling can offer insights into mechanisms of action of new candidate drugs that emerge from screens. For instance, the transcriptional signature of cell wall disruptors (e.g., β-lactams) is distinct from that of DNA-damaging agents (e.g., fluoroquinolones) or protein synthesis inhibitors (e.g., aminoglycosides). Thus, transcriptional analysis of a pathogen’s response to a new antibiotic can either suggest a mechanism of action or flag compounds for prioritization because of a potentially novel activity. In an alternative genomic strategy for determining mechanisms of action, an RNA interference approach followed by targeted sequencing has been used to identify genes required for antitrypanosomal drug efficacy. This approach provided new insights into the mechanism of action of drugs that have been in use for decades for human African trypanosomiasis. Third, sequencing can readily identify the most conserved regions of a pathogen’s genomes and corresponding gene products; this information is invaluable in narrowing antigen candidates for vaccine development. These surface proteins can be expressed recombinantly and tested for the ability to elicit a serologic response and protective immunity. This process, termed reverse vaccinology, has proved particularly useful for pathogens that are difficult to culture or poorly immunogenic, as was the case with the development of a vaccine for Neisseria meningitidis serogroup B.
Large-scale gene content analysis from sequencing or expression profiling enables new research directions that provide novel insights into the interplay of pathogen and host during infection or colonization. One important goal of such research is to suggest new therapeutic approaches to disrupt this interaction in favor of the host. Indeed, one of the most immediate applications of next-generation sequencing technology has come from simply characterizing human pathogens and related commensal or environmental strains and then finding genomic correlates for pathogenicity. For instance, as Escherichia coli varies from a simple nonpathogenic, lab-adapted strain (K-12) to a Shiga toxin–producing enterohemorrhagic gastrointestinal pathogen (O157:H7), it displays up to a 25% difference in gene content, even though its phylogenetic classification stays the same. Although this is an extreme example, it is not an isolated case. Some isolates of Enterococcus—notorious for its increasing incidence of resistance to common antibiotics such as ampicillin, vancomycin, and aminoglycosides—also contain recently acquired genetic material comprising up to 25% of the genome on mobile genetic elements. This fact suggests that horizontal gene transfer may play an important role in the organism’s adaptation as a nosocomial pathogen. On closer study, this genome expansion has been demonstrated to be associated with loss of regulatory elements called CRISPRs (clustered, regularly interspaced short palindromic repeats). Loss of CRISPR elements, which protect the bacterial genome from invasion by certain foreign genetic materials, may thus facilitate the acquisition of antibiotic resistance–conferring genetic elements. While loss of this regulation appears to impose a competitive disadvantage in antibiotic-free environments, these drug-resistant strains thrive in the presence of even some of the most useful antienterococcal therapies. In addition to insights gained from genome sequencing, extension of unbiased whole-transcriptome sequencing (RNA-Seq) efforts to bacteria is beginning to identify unexpected regulatory, noncoding RNAs in many diverse species. While the functional implications of these new transcripts are as yet largely unknown, the presence of such features—conserved across many bacterial species—implies evolutionary importance and suggests areas for future study and possible new therapeutic avenues.
Thus, genomic studies are already beginning to transform our understanding of infection, offering evidence of virulence factors or toxins and providing insight into ongoing evolution of pathogenicity and drug resistance. One goal of such studies is to identify therapeutic agents that can disrupt the pathogenic process; there is currently much interest in the theoretical concept of antivirulence drugs that inhibit virulence factors rather than killing the pathogen outright as a means to intervene in infection. Further, as sequencing becomes increasingly accessible and efficient, large-scale studies with unprecedented statistical power to associate clinical outcomes with pathogen and host genotypes and thus to further reveal vulnerabilities in the infection process that can be targeted for disruption are being initiated. Although this is just the beginning, such studies point to a tantalizing future in which the clinician is armed with genomic predictors of infection outcome and therapeutic response to guide clinical decision-making.
EPIDEMIOLOGY OF INFECTIOUS DISEASES
Epidemiologic studies of infectious diseases have several main goals: to identify and characterize outbreaks, to describe the pattern and dynamics of an infectious disease as it spreads through populations, and to identify interventions that can limit or reduce the burden of disease. One classic, paradigmatic example is John Snow’s elucidation of the origin of the 1854 London cholera outbreak. Snow used careful geographic mapping of cases to determine that the likely source of the outbreak was contaminated water from the Broad Street pump, and, by removing the pump handle, he aborted the outbreak. Whereas that intervention was undertaken without knowledge of the causative agent of cholera, advances in microbiology and genomics have expanded the purview of epidemiology, which now considers not just the disease but also the pathogen, its virulence factors, and the complex relationships between microbial and host populations.
Through the use of novel genomic tools such as high-throughput sequencing, the diversity of a microbial population can now be rapidly described with unprecedented resolution, with discrimination between isolates that have single-nucleotide differences across the entire genome and advancement beyond prior approaches that relied on phenotypes (such as antibiotic resistance testing) or genetic markers (such as multilocus sequence typing). The development of statistical methods grounded in molecular genetics and evolutionary theory has established analytical approaches that translate descriptions of microbial population diversity and structure into insights into the origin and history of pathogen spread. By linking phylogenetic reconstruction with epidemiologic and demographic data, genomic epidemiology provides the opportunity to track transmission from person to person, to infer transmission patterns of both pathogens and sequence elements that confer phenotypes of interest, and to estimate the transmission dynamics of outbreaks.
DECIPHERING PERSON-TO-PERSON TRANSMISSION
The use of comparisons of whole-genome sequencing to infer person-to-person transmission and identify point-source outbreaks of pathogens has proved useful in hospital infection control settings. As reported in a seminal paper in 2010, a study of MRSA in a Thai hospital demonstrated that whole-genome sequencing can be used to infer transmission of a pathogen from patient to patient within a hospital setting through integration of the analysis of accumulation of mutations over time with dates and hospital locations of the infections. Since that time, multiple instances of the use of whole-genome sequencing to define and motivate interventions aimed at interrupting transmission chains have been reported. In another MRSA outbreak in a special-care baby unit in Cambridge, United Kingdom, whole-genome sequencing extended the traditional infection control analysis, which relies on typing organisms by their antibiotic susceptibilities, to sequencing of isolates from clinical samples. This approach identified an otherwise unrecognized outbreak of a specific MRSA strain that was occurring against a background of the usual pattern of infections caused by a diverse circulating population of MRSA strains. The analysis showed evidence of transmission among mothers within the special-care baby unit and in the community and demonstrated the key role of MRSA carriage in a single health care provider in the persistence of the outbreak. MRSA decolonization of the health care provider terminated the outbreak. In yet another example, in response to the observation of 18 cases of infection by carbapenemase-producing Klebsiella pneumoniae over 6 months at the National Institutes of Health Clinical Research Center, genome sequencing of the isolates was used to discriminate between the possibilities that these cases represented multiple, independent introductions into the health care system or a single introduction with subsequent transmission. On the basis of network and phylogenetic analysis of genomic and epidemiologic data, the authors reconstructed the likely relationships among the isolates from patient to patient, demonstrating that the spread of resistant Klebsiella infection was in fact due to nosocomial transmission of a single strain.
Uncovering of unexpected transmission events by genomic epidemiology studies is motivating renewed questioning of pathogen ecology and modes of transmission. For example, the rise in prevalence of infections with nontuberculous mycobacteria, including Mycobacterium abscessus, among patients with cystic fibrosis (CF) has led to speculation about the possible role of patient-to-patient transmission in the CF community; however, conventional typing approaches have lacked the resolution to define population structure accurately, a critical component of inferring transmission. Past infection control guidelines discounted the possibility of acquisition of nontuberculous mycobacteria in health care settings, as no strong evidence for such transmission had been described. In a whole-genome sequencing study of M. abscessus isolates from patients with CF, an analytical approach using genome sequencing, epidemiology, and Bayesian modeling examined the likelihood of transmission between patients within a CF center; the authors found nearly identical isolates in a number of patients and observed that these isolates were less diverse than isolates from a single individual. Because no clear epidemiologic link places the infected patients in the same place at the same time, this finding highlights a need to explore preexisting notions of circumstances required for transmission and a reconsideration of M. abscessus infection control guidelines. Similar studies of other pathogens—particularly those that share human, other animal host, and environmental reservoirs—will continue to advance our insight into the relative roles and prominence of sources of infection as well as the modes of spread through populations, thereby establishing evidence-based strategies for prevention and intervention.
As increasing numbers of studies aim to carefully define the origins and spread of infectious agents using the high-resolution lens of whole-genome sequencing, fundamental questions are arising with regard to our understanding of infection in a single individual and the process of a single transmission event. For example, a better understanding of a pathogen population’s diversity within a single infected individual is a critical component in interpreting the relationship among isolates from different patients. While we have traditionally thought of individuals as infected with a single bacterial strain, a recent sequencing study of multiple colonies of S. aureus from a single individual showed a “cloud” of diversity; this finding raises a number of questions that will be important to address as this field develops: What is the clinical significance of this diversity? What are the processes that generate and limit diversity? What amount of diversity is transmitted under different conditions and routes of transmission? How do the answers to these questions vary by infectious organism, type of infection, and host and in response to treatment? More comprehensive descriptions of diversity, population dynamics, transmission bottlenecks, and the forces that shape and influence the growth and spread of microbial populations will be a critically important focus of future investigations.
RECONSTRUCTING THE ORIGINS AND DYNAMICS OF PATHOGEN SPREAD
In addition to reconstructing the transmission chains of local outbreaks, genomics-based epidemiologic methods are providing insight into broad-scale geographic and temporal spread of pathogens. A classic example has been the study of cholera, the dehydrating diarrheal illness caused by infection with Vibrio cholerae. Cholera first spread worldwide from the Indian subcontinent in the 1800s and has since caused seven pandemics; the seventh pandemic has been ongoing since the 1960s. An investigation into the geographic patterns of cholera spread in the seventh pandemic used genome sequences from a global collection of 154 V. cholerae strains representing isolates from 1957–2010. This investigation revealed that the seventh pandemic has comprised at least three overlapping waves spreading out from the Indian subcontinent (Fig. 146-4). Further, analysis of the genome of an isolate of V. cholerae from the 2010 outbreak of cholera in Haiti showed it to be more closely related to isolates from South Asia than to isolates from neighboring Latin America, a result supporting the hypothesis that the outbreak was derived from V. cholerae introduced into Haiti by human travel (likely from Nepal) rather than by environmental or more geographically proximal sources. A subsequent study that dated the time to the most recent common ancestor of a population of V. cholerae isolates from Haiti provided further support for a single point-source introduction from Nepal.
FIGURE 146-4 Transmission events inferred from phylogenetic reconstruction of 154 Vibrio cholerae isolates from the seventh cholera pandemic. Date ranges represent estimated time to the most recent common ancestor for strains transmitted from source to destination locations, based on a Bayesian model of the phylogeny. (Reprinted with permission from A Mutreja et al: Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477:462, 2011.)
Increasing numbers of investigations into the spread of many pathogens—thus far including strains of S. aureus, S. pneumoniae, Chlamydia, Salmonella, Shigella, E. coli, C. difficile, West Nile virus, rabies virus, and dengue virus—are contributing to a growing atlas of maps describing routes, patterns, and tempos of microbial diversification and dissemination. Large-scale efforts like the 100K Foodborne Pathogen Genome Project, which aims to sequence the genomes of 100,000 strains of food-borne pathogens collected from sources including food, the environment, and farm animals, are possible because of advances in sequencing technologies. Such studies will yield a vast amount of data that can be used to investigate diversity and microbiologic links within distinct niches and the patterns of spread from one niche to another. The increasingly broad adoption of genome sequencing by health care and public health institutions will ensure that the available catalog of genome sequences and associated epidemiologic data will grow very rapidly. With higher-resolution description of microbial diversity and of the dynamics of that diversity over time and across epidemiologic and demographic boundaries and evolutionary niches, we will gain even greater insights into the relationships of transmission routes and patterns of historic spread.
PREDICTING EPIDEMIC POTENTIAL
Defining pathogen transmissibility is a critical step in the development of public health surveillance and intervention strategies as this information can help predict the epidemic potential of an outbreak. Transmissibility can be estimated by a variety of methods, including inference from the growth rate of an epidemic together with the generation time of an infection (the mean interval between infection of an index case and of the people infected by that index case). Genome sequencing and analysis of a well-sampled population provide another method by which to derive similar fundamental epidemiologic parameters. One key measure of transmissibility is the basic reproduction number (R0), defined as the number of secondary infections generated from a single primary infectious case. When the basic reproduction number is greater than 1 (R0 >1), an outbreak has epidemic potential; when it is less than 1 (R0 <1), the outbreak will become extinct. On the basis of sequences from influenza samples obtained from infected patients very early in the 2009 H1N1 influenza pandemic, the basic reproduction number was estimated through a population genomic analysis at 1.2; this compared to estimates of 1.4–1.6 based on several epidemiologic analyses. In addition, with the assumption of a molecular clock model, sequences of H1N1 samples together with information about when and where the samples were obtained have been used to estimate the date and location of the pandemic’s origin, providing insight into disease origins and dynamics. Because the magnitude and intensity of the public health response are guided by the predicted size of an outbreak, the ability of genomic methods to elucidate a pathogen’s origin and epidemic potential adds an important dimension to the contributions of these methods to infectious disease epidemiology.
PROVIDING INSIGHT INTO PATHOGEN EVOLUTION
Beyond describing transmission and dynamics, pathogen genomics can provide insight into the evolution of pathogens and the interaction of selective pressures, the host, and pathogen populations, which can have implications for vaccine or therapeutic development. From a clinical perspective, this process is central to the acquisition of antibiotic resistance, the generation of increasing pathogenicity or new virulence traits, the evasion of host immunity and clearance (leading to chronic infection), and vaccine efficacy.
Microbial genomes evolve through a variety of mechanisms, including mutation, duplication, insertion, deletion, recombination, and horizontal gene transfer. Segmented viruses (e.g., influenza virus) can reassort gene segments within multiply infected cells. The pandemic 2009 H1N1 influenza A virus, for example, appears to have been generated through reassortment of several avian, swine, and human influenza strains. Such potential for the evolution of novel pandemic strains has precipitated concern about the possible evolution to transmissibility of virulent strains that have been associated with high mortality rates but have not yet exhibited efficient human infectivity. Controversial experiments with H5N1 avian influenza virus, for example, have defined five mutations that render the virus transmissible, at least in ferrets—the animal model system for human influenza.
The continual antigenic evolution of seasonal influenza offers an example of how studies of pathogen evolution can impact surveillance and vaccine development. Frequent updates to the annual influenza vaccine are needed to ensure protection against the dominant strains. These updates are based on an ability to anticipate which viral populations from a pool of substantial locally and globally diverse circulating viruses will predominate in the upcoming season. Toward that end, sequencing-based studies of influenza virus dynamics have shed light on the global spread of influenza, providing concrete data on patterns of spread and helping to elucidate the origins, emergence, and circulation of novel strains. Through analysis of more than 1000 influenza A H3N2 virus isolates over the 2002–2007 influenza seasons, Southeast Asia was identified as the usual site from which diversity originates and spreads worldwide. Further studies of global isolate collections have shed further light on the diversity of circulating virus, showing that some strains persist and circulate outside of Asia for multiple seasons.
Not only do genomic epidemiology studies have the potential to help guide vaccine selection and development; they are also helping to track what happens to pathogens circulating in the population in response to vaccination. By describing pathogen evolution under the selective pressure of a vaccinated population, such studies can play a key role in surveillance and identification of virulence determinants and perhaps may even help to predict the future evolution of escape from vaccine protection. The 7-valent pneumococcal conjugate vaccine (PCV-7) targeted the seven serotypes of S. pneumoniae responsible for the majority of cases of invasive disease at the time of its introduction in 2000; since then, PCV-7 has dramatically reduced the incidence of pneumococcal disease and mortality. Population genomic analysis of the sequences of more than 600 Massachusetts pneumococcal isolates from 2001–2007 has shown that preexisting rare nonvaccine serotypes are replacing vaccine serotypes and that some strains have persisted despite vaccination by recombining the vaccine-targeted capsule locus with a cassette of capsule genes from non-vaccine-targeted serotypes.
GLOBAL CONSIDERATIONS
![]() While cutting-edge genomic technologies are largely implemented in the developed world, their application to infectious diseases offers perhaps the biggest potential impact in less developed regions where the burden of these infections is greatest. This globalization of genomic technology and its extensions has already begun in each of the areas of focus highlighted in this chapter; it has occurred both through the application of advanced technologies to samples collected in the developing world and through the adaptation and importation of technologies directly to the developing world for on-site implementation as they become more globally accessible. Genomic characterization of the pathogens responsible for important global illnesses such as tuberculosis, malaria, trypanosomiasis, and cholera has led to insights in diagnosis, treatment, and infection control. For instance, the nucleic acid–based test developed for rapid diagnosis of M. tuberculosis infection and detection of rifampin resistance is being priced for implementation in field settings in Africa and Asia where tuberculosis is most prevalent. The potential to diagnose multidrug-resistant tuberculosis in hours instead of weeks or months may truly revolutionize treatment and control of this common and devastating illness. High-resolution genomic tracking of the spread of cholera has yielded insights into which public health measures may prove most effective in controlling local epidemics. Overall, sequencing efforts have become exponentially cheaper with each passing year. As these technologies synergize with efforts to globalize information-technology resources, global implementation of genomic methods promises to spread state-of-the-art methods for diagnosis, treatment, and epidemic tracking of infections to areas that need these capabilities the most.
While cutting-edge genomic technologies are largely implemented in the developed world, their application to infectious diseases offers perhaps the biggest potential impact in less developed regions where the burden of these infections is greatest. This globalization of genomic technology and its extensions has already begun in each of the areas of focus highlighted in this chapter; it has occurred both through the application of advanced technologies to samples collected in the developing world and through the adaptation and importation of technologies directly to the developing world for on-site implementation as they become more globally accessible. Genomic characterization of the pathogens responsible for important global illnesses such as tuberculosis, malaria, trypanosomiasis, and cholera has led to insights in diagnosis, treatment, and infection control. For instance, the nucleic acid–based test developed for rapid diagnosis of M. tuberculosis infection and detection of rifampin resistance is being priced for implementation in field settings in Africa and Asia where tuberculosis is most prevalent. The potential to diagnose multidrug-resistant tuberculosis in hours instead of weeks or months may truly revolutionize treatment and control of this common and devastating illness. High-resolution genomic tracking of the spread of cholera has yielded insights into which public health measures may prove most effective in controlling local epidemics. Overall, sequencing efforts have become exponentially cheaper with each passing year. As these technologies synergize with efforts to globalize information-technology resources, global implementation of genomic methods promises to spread state-of-the-art methods for diagnosis, treatment, and epidemic tracking of infections to areas that need these capabilities the most.
SUMMARY
By illuminating the genetic information that encodes the most fundamental processes of life, genomic technologies are transforming many aspects of medicine. In infectious diseases, methods such as next-generation sequencing and genome-scale expression analysis offer information of unprecedented depth about individual microbes as well as microbial communities. This information is expanding our understanding of the interactions of these microorganisms and communities with one another, with their human hosts, and with the environment. Despite significant progress and the abundant genomic data now available, technological and financial barriers continue to impede the widespread adoption of large-scale pathogen sequencing in clinical, public health, and research settings. As even vaster amounts of data are generated, innovations in storage, development of bioinformatics tools to manipulate the data, standardization of methods, and training of end-users in both the research and clinical realms will be required. The cost-effectiveness and applicability of whole-genome sequencing, particularly in the clinic, remain to be studied, and studies of the impact of genome sequencing on patient outcomes will be needed to clarify the contexts in which these new methodologies can make the greatest contributions to patient well-being. The ongoing efforts to overcome limitations through collaboration, teaching, and reduction of financial obstacles should be applauded and expanded. With advances in genomic technologies and computational analysis, our ability to detect, characterize, treat, monitor, prevent, and control infections has progressed rapidly in recent years and will continue to do so, with the hope of heralding a new era where the clinician is better armed to combat infection and promote human health.
147 |
Approach to the Acutely Ill Infected Febrile Patient |
The physician treating the acutely ill febrile patient must be able to recognize infections that require emergent attention. If such infections are not adequately evaluated and treated at initial presentation, the opportunity to alter an adverse outcome may be lost. In this chapter, the clinical presentations of and approach to patients with relatively common infectious disease emergencies are discussed. These infectious processes and their treatments are discussed in detail in other chapters.
|
TREATMENT |
THE ACUTELY ILL PATIENT |
In the acutely ill patient, empirical antibiotic therapy is critical and should be administered without undue delay. Increased prevalence of antibiotic resistance in community-acquired bacteria must be considered when antibiotics are selected. Table 147-1 lists first-line empirical regimens for infections considered in this chapter. In addition to the rapid initiation of antibiotic therapy, several of these infections require urgent surgical attention. Neurosurgical evaluation for subdural empyema, otolaryngologic surgery for possible mucormycosis, and cardiothoracic surgery for critically ill patients with acute endocarditis are as important as antibiotic therapy. For infections such as necrotizing fasciitis and clostridial myonecrosis, rapid surgical intervention supersedes other diagnostic or therapeutic maneuvers.
|
EMPIRICAL TREATMENT FOR COMMON INFECTIOUS DISEASE EMERGENCIESa |
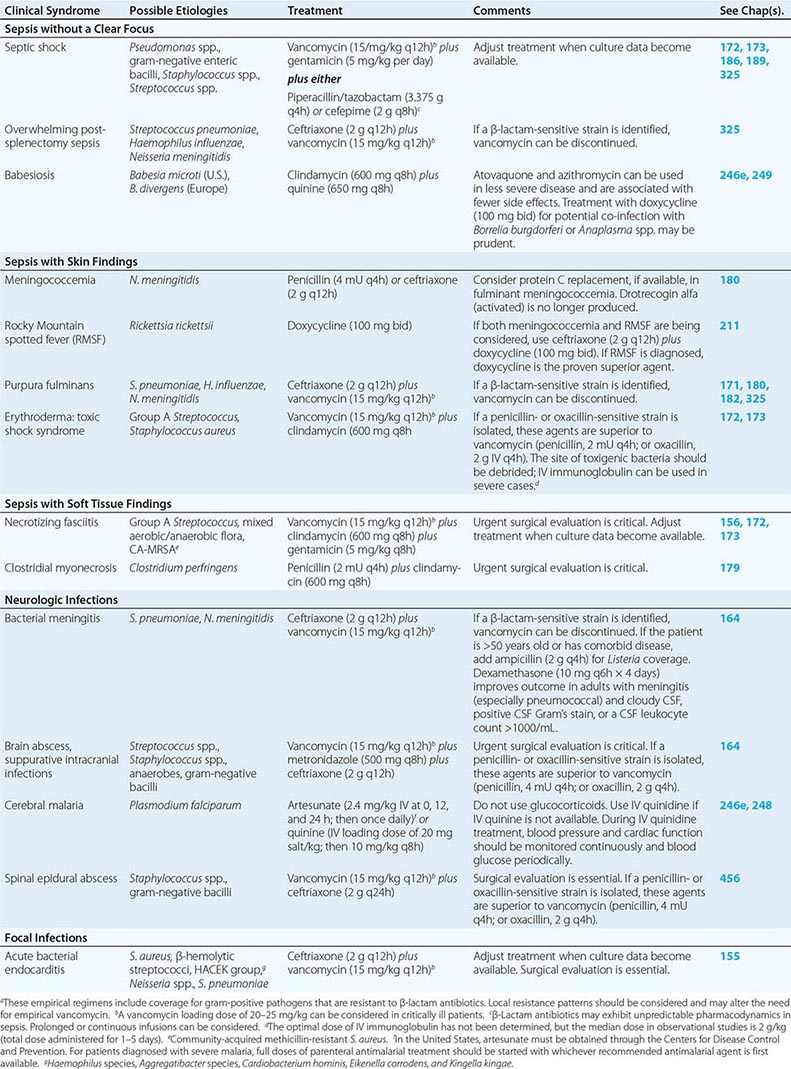
Adjunctive treatments may reduce morbidity and mortality rates and include dexamethasone for bacterial meningitis or IV immunoglobulin for TSS and necrotizing fasciitis caused by group A Streptococcus. Adjunctive therapies should usually be initiated within the first hours of treatment; however, dexamethasone for bacterial meningitis must be given before or at the time of the first dose of antibiotic. Glucocorticoids can also be harmful, sometimes resulting in worse outcomes—e.g., when given in the setting of cerebral malaria or viral hepatitis.
SPECIFIC PRESENTATIONS
The infections considered below according to common clinical presentation can have rapidly catastrophic outcomes, and their immediate recognition and treatment can be life-saving. Recommended empirical therapeutic regimens are presented in Table 147-1.
SEPSIS WITHOUT AN OBVIOUS FOCUS OF PRIMARY INFECTION
Patients initially have a brief prodrome of nonspecific symptoms and signs that progresses quickly to hemodynamic instability with hypotension, tachycardia, tachypnea, respiratory distress, and altered mental status. Disseminated intravascular coagulation (DIC) with clinical evidence of a hemorrhagic diathesis is a poor prognostic sign.
Septic Shock (See also Chap. 325) Patients with bacteremia leading to septic shock may have a primary site of infection (e.g., pneumonia, pyelonephritis, or cholangitis) that is not evident initially. Elderly patients with comorbid conditions, hosts compromised by malignancy and neutropenia, and patients who have recently undergone a surgical procedure or hospitalization are at increased risk for an adverse outcome. Gram-negative bacteremia with organisms such as Pseudomonas aeruginosa or Escherichia coli and gram-positive infection with organisms such as Staphylococcus aureus (including methicillin-resistant S. aureus [MRSA]) or group A streptococci can present as intractable hypotension and multiorgan failure. Treatment can usually be initiated empirically on the basis of the presentation, host factors (Chap. 325), and local patterns of bacterial resistance. Outcomes are worse when antimicrobial treatment is delayed or when the responsible pathogen ultimately proves not to be susceptible to the initial regimen. Broad-spectrum antimicrobial agents are therefore recommended and should be instituted rapidly, preferably within the first hour after presentation. Risk factors for fungal infection should be assessed, as the incidence of fungal septic shock is increasing. Biomarkers such as C-reactive protein and procalcitonin have not proved reliable diagnostically but, when measured over time, can facilitate appropriate de-escalation of therapy. Glucocorticoids should be considered only for patients with severe sepsis who do not respond to fluid resuscitation and vasopressor therapy.
Overwhelming Infection in Asplenic Patients (See also Chap. 325) Patients without splenic function are at risk for overwhelming bacterial sepsis. Asplenic adult patients succumb to sepsis at 58 times the rate of the general population. Most infections are thought to occur within the first 2 years after splenectomy, with a mortality rate of ~50%, but the increased risk persists throughout life. In asplenia, encapsulated bacteria cause the majority of infections. Adults, who are more likely to have antibody to these organisms, are at lower risk than children. Streptococcus pneumoniae is the most common isolate, causing 50–70% of cases, but the risk of infection with Haemophilus influenzae or Neisseria meningitidis is also high. Severe clinical manifestations of infections due to E. coli, S. aureus, group B streptococci, P. aeruginosa, Bordetella holmesii, and Capnocytophaga, Babesia, and Plasmodium species have been described.
Babesiosis (See also Chap. 249) A history of recent travel to endemic areas raises the possibility of infection with Babesia. Between 1 and 4 weeks after a tick bite, the patient experiences chills, fatigue, anorexia, myalgia, arthralgia, shortness of breath, nausea, and headache; ecchymosis and/or petechiae are occasionally seen. The tick that most commonly transmits Babesia, Ixodes scapularis, also transmits Borrelia burgdorferi (the agent of Lyme disease) and Anaplasma; co-infection can occur, resulting in more severe disease. Infection with the European species Babesia divergens is more frequently fulminant than that due to the U.S. species Babesia microti. B. divergens causes a febrile syndrome with hemolysis, jaundice, hemoglobinemia, and renal failure and is associated with a mortality rate of >40%. Severe babesiosis is especially common in asplenic hosts but does occur in hosts with normal splenic function, particularly those >60 years of age and those with underlying immunosuppressive conditions such as HIV infection or malignancy. Complications include renal failure, acute respiratory failure, and DIC.
Other Sepsis Syndromes Tularemia (Chap. 195) is seen throughout the United States but occurs primarily in Arkansas, Missouri, South Dakota, and Oklahoma. This disease is associated with wild rabbit, tick, and tabanid fly contact. It can be transmitted by arthropod bite, handling of infected animal carcasses, consumption of contaminated food and water, or inhalation. The typhoidal form can be associated with gram-negative septic shock and a mortality rate of >30%, especially in patients with underlying comorbid or immunosuppressive conditions. Plague occurs infrequently in the United States (Chap. 196), primarily after contact with ground squirrels, prairie dogs, or chipmunks, but is endemic in other parts of the world, with >90% of all cases occurring in Africa. The septic form is particularly rare and is associated with shock, multiorgan failure, and a 30% mortality rate. These infections should be considered in the appropriate epidemiologic setting. The Centers for Disease Control and Prevention lists Francisella tularensis and Yersinia pestis (the agents of tularemia and plague, respectively) along with Bacillus anthracis (the agent of anthrax) as important organisms that might be used for bioterrorism (Chap. 261e).
SEPSIS WITH SKIN MANIFESTATIONS
(See also Chap. 24) Maculopapular rashes may reflect early meningococcal or rickettsial disease but are usually associated with nonemergent infections. Exanthems are usually viral. Primary HIV infection commonly presents with a rash that is typically maculopapular and involves the upper part of the body but can spread to the palms and soles. The patient is usually febrile and can have lymphadenopathy, severe headache, dysphagia, diarrhea, myalgias, and arthralgias. Recognition of this syndrome provides an opportunity to prevent transmission and to institute treatment and monitoring early on.
Petechial rashes caused by viruses are seldom associated with hypotension or a toxic appearance, although there can be exceptions (e.g., severe measles or arboviral infection). Petechial rashes limited to the distribution of the superior vena cava are rarely associated with severe disease. In other settings, petechial rashes require more urgent attention.
![]() Meningococcemia (See also Chap. 180) Almost three-quarters of patients with N. meningitidis bacteremia have a rash. Meningococcemia most often affects young children (i.e., those 6 months to 5 years old). In sub-Saharan Africa, the high prevalence of serogroup A meningococcal disease has been a threat to public health for more than a century. Thousands of deaths occur annually in this area, which is known as the “meningitis belt,” and large epidemic waves occur approximately every 8–12 years. Serogroups W135 and × are also important emerging pathogens in Africa. In the United States, sporadic cases and outbreaks occur in day-care centers, schools (grade school through college, particularly among college freshmen living in residential halls), and army barracks. Household contacts of index cases are at 400–800 times greater risk of disease than the general population. Patients may exhibit fever, headache, nausea, vomiting, myalgias, changes in mental status, and meningismus. However, the rapidly progressive form of disease is not usually associated with meningitis. The rash is initially pink, blanching, and maculopapular, appearing on the trunk and extremities, but then becomes hemorrhagic, forming petechiae. Petechiae are first seen at the ankles, wrists, axillae, mucosal surfaces, and palpebral and bulbar conjunctiva, with subsequent spread on the lower extremities and to the trunk. A cluster of petechiae may be seen at pressure points—e.g., where a blood pressure cuff has been inflated. In rapidly progressive meningococcemia (10–20% of cases), the petechial rash quickly becomes purpuric (see Fig. 70-5), and patients develop DIC, multiorgan failure, and shock; 50–60% of these patients die, and survivors often require extensive debridement or amputation of gangrenous extremities. Hypotension with petechiae for <12 h is associated with significant mortality. Cyanosis, coma, oliguria, metabolic acidosis, and elevated partial thromboplastin time also are associated with a fatal outcome. Correction of protein C deficiency may improve outcome. Antibiotics given in the office by the primary care provider before hospital evaluation and admission may improve prognosis; this observation suggests that early initiation of treatment may be life-saving. Meningococcal conjugate vaccines are protective against serogroups A, C, Y and W135 and are recommended for children 11–18 years of age and for other high-risk patients.
Meningococcemia (See also Chap. 180) Almost three-quarters of patients with N. meningitidis bacteremia have a rash. Meningococcemia most often affects young children (i.e., those 6 months to 5 years old). In sub-Saharan Africa, the high prevalence of serogroup A meningococcal disease has been a threat to public health for more than a century. Thousands of deaths occur annually in this area, which is known as the “meningitis belt,” and large epidemic waves occur approximately every 8–12 years. Serogroups W135 and × are also important emerging pathogens in Africa. In the United States, sporadic cases and outbreaks occur in day-care centers, schools (grade school through college, particularly among college freshmen living in residential halls), and army barracks. Household contacts of index cases are at 400–800 times greater risk of disease than the general population. Patients may exhibit fever, headache, nausea, vomiting, myalgias, changes in mental status, and meningismus. However, the rapidly progressive form of disease is not usually associated with meningitis. The rash is initially pink, blanching, and maculopapular, appearing on the trunk and extremities, but then becomes hemorrhagic, forming petechiae. Petechiae are first seen at the ankles, wrists, axillae, mucosal surfaces, and palpebral and bulbar conjunctiva, with subsequent spread on the lower extremities and to the trunk. A cluster of petechiae may be seen at pressure points—e.g., where a blood pressure cuff has been inflated. In rapidly progressive meningococcemia (10–20% of cases), the petechial rash quickly becomes purpuric (see Fig. 70-5), and patients develop DIC, multiorgan failure, and shock; 50–60% of these patients die, and survivors often require extensive debridement or amputation of gangrenous extremities. Hypotension with petechiae for <12 h is associated with significant mortality. Cyanosis, coma, oliguria, metabolic acidosis, and elevated partial thromboplastin time also are associated with a fatal outcome. Correction of protein C deficiency may improve outcome. Antibiotics given in the office by the primary care provider before hospital evaluation and admission may improve prognosis; this observation suggests that early initiation of treatment may be life-saving. Meningococcal conjugate vaccines are protective against serogroups A, C, Y and W135 and are recommended for children 11–18 years of age and for other high-risk patients.
Rocky Mountain Spotted Fever (See also Chap. 211) RMSF is a tickborne disease caused by Rickettsia rickettsii that occurs throughout North and South America. Up to 40% of patients do not report a history of a tick bite, but a history of travel or outdoor activity (e.g., camping in tick-infested areas) can often be ascertained. For the first 3 days, headache, fever, malaise, myalgias, nausea, vomiting, and anorexia are documented. By day 3, half of patients have skin findings. Blanching macules develop initially on the wrists and ankles and then spread over the legs and trunk. The lesions become hemorrhagic and are frequently petechial. The rash spreads to palms and soles later in the course. The centripetal spread is a classic feature of RMSF but occurs in a minority of patients. Moreover, 10–15% of patients with RMSF never develop a rash. The patient can be hypotensive and develop noncardiogenic pulmonary edema, confusion, lethargy, and encephalitis progressing to coma. The CSF contains 10–100 cells/μL, usually with a predominance of mononuclear cells. The CSF glucose level is often normal; the protein concentration may be slightly elevated. Renal and hepatic injury as well as bleeding secondary to vascular damage are noted. For untreated infections, mortality rates are 20–30%. Delayed recognition and treatment are associated with a greater risk of death; Native Americans, children 5–9 years of age, adults >70 years old, and persons with underlying immunosuppression also are at increased risk of death.
![]() Other rickettsial diseases cause significant morbidity and mortality worldwide. Mediterranean spotted fever caused by Rickettsia conorii is found in Africa, southwestern and south-central Asia, and southern Europe. Patients have fever, flu-like symptoms, and an inoculation eschar at the site of the tick bite. A maculopapular rash develops within 1–7 days, involving the palms and soles but sparing the face. Elderly patients or those with diabetes, alcoholism, uremia, or congestive heart failure are at risk for severe disease characterized by neurologic involvement, respiratory distress, and gangrene of the digits. Mortality rates associated with this severe form of disease approach 50%. Epidemic typhus, caused by Rickettsia prowazekii, is transmitted in louse-infested environments and emerges in conditions of extreme poverty, war, and natural disaster. Patients experience a sudden onset of high fevers, severe headache, cough, myalgias, and abdominal pain. A maculopapular rash develops (primarily on the trunk) in more than half of patients and can progress to petechiae and purpura. Serious signs include delirium, coma, seizures, noncardiogenic pulmonary edema, skin necrosis, and peripheral gangrene. Mortality rates approached 60% in the preantibiotic era and continue to exceed 10–15% in contemporary outbreaks. Scrub typhus, caused by Orientia tsutsugamushi (a separate genus in the family Rickettsiaceae), is transmitted by larval mites or chiggers and is one of the most common infections in southeastern Asia and the western Pacific. The organism is found in areas of heavy scrub vegetation (e.g., along riverbanks). Patients may have an inoculation eschar and may develop a maculopapular rash. Severe cases progress to pneumonia, meningoencephalitis, DIC, and renal failure. Mortality rates range from 1% to 35%.
Other rickettsial diseases cause significant morbidity and mortality worldwide. Mediterranean spotted fever caused by Rickettsia conorii is found in Africa, southwestern and south-central Asia, and southern Europe. Patients have fever, flu-like symptoms, and an inoculation eschar at the site of the tick bite. A maculopapular rash develops within 1–7 days, involving the palms and soles but sparing the face. Elderly patients or those with diabetes, alcoholism, uremia, or congestive heart failure are at risk for severe disease characterized by neurologic involvement, respiratory distress, and gangrene of the digits. Mortality rates associated with this severe form of disease approach 50%. Epidemic typhus, caused by Rickettsia prowazekii, is transmitted in louse-infested environments and emerges in conditions of extreme poverty, war, and natural disaster. Patients experience a sudden onset of high fevers, severe headache, cough, myalgias, and abdominal pain. A maculopapular rash develops (primarily on the trunk) in more than half of patients and can progress to petechiae and purpura. Serious signs include delirium, coma, seizures, noncardiogenic pulmonary edema, skin necrosis, and peripheral gangrene. Mortality rates approached 60% in the preantibiotic era and continue to exceed 10–15% in contemporary outbreaks. Scrub typhus, caused by Orientia tsutsugamushi (a separate genus in the family Rickettsiaceae), is transmitted by larval mites or chiggers and is one of the most common infections in southeastern Asia and the western Pacific. The organism is found in areas of heavy scrub vegetation (e.g., along riverbanks). Patients may have an inoculation eschar and may develop a maculopapular rash. Severe cases progress to pneumonia, meningoencephalitis, DIC, and renal failure. Mortality rates range from 1% to 35%.
If recognized in a timely fashion, rickettsial disease is very responsive to treatment. Doxycycline (100 mg twice daily for 3–14 days) is the treatment of choice for both adults and children. The newer macrolides and chloramphenicol may be suitable alternatives, but mortality rates are higher when a tetracycline-based treatment is not given.
Purpura Fulminans (See also Chaps. 180 and 325) Purpura fulminans is the cutaneous manifestation of DIC and presents as large ecchymotic areas and hemorrhagic bullae. Progression of petechiae to purpura, ecchymoses, and gangrene is associated with congestive heart failure, septic shock, acute renal failure, acidosis, hypoxia, hypotension, and death. Purpura fulminans has been associated primarily with N. meningitidis but, in splenectomized patients, may be associated with S. pneumoniae, H. influenzae, and S. aureus.
Ecthyma Gangrenosum Septic shock caused by P. aeruginosa or Aeromonas hydrophila can be associated with ecthyma gangrenosum (see Figs. 189-1 and 25e-35): hemorrhagic vesicles surrounded by a rim of erythema with central necrosis and ulceration. These gram-negative bacteremias are most common among patients with neutropenia, extensive burns, and hypogammaglobulinemia.
Other Emergent Infections Associated with Rash Vibrio vulnificus and other noncholera Vibrio bacteremic infections (Chap. 193) can cause focal skin lesions and overwhelming sepsis in hosts with chronic liver disease, iron storage disorders, diabetes, renal insufficiency, or other immunocompromising conditions. After ingestion of contaminated raw shellfish, typically oysters from the Gulf Coast, there is a sudden onset of malaise, chills, fever, and hypotension. The patient develops bullous or hemorrhagic skin lesions, usually on the lower extremities, and 75% of patients have leg pain. The mortality rate can be as high as 50–60%, particularly when the patient presents with hypotension. Outcomes are improved when patients are treated with tetracycline-containing regimens. Other infections, caused by agents such as Aeromonas, Klebsiella, and E. coli, can cause hemorrhagic bullae and death due to overwhelming sepsis in cirrhotic patients. Capnocytophaga canimorsus can cause septic shock in asplenic patients. Infection typically follows a dog bite. Patients present with fever, chills, myalgia, vomiting, diarrhea, dyspnea, confusion, and headache. Findings can include an exanthem or erythema multiforme (see Figs. 70-9 and 25e-25), cyanotic mottling or peripheral cyanosis, petechiae, and ecchymosis. About 30% of patients with this fulminant form die of overwhelming sepsis and DIC, and survivors may require amputation because of gangrene.
Erythroderma TSS (Chaps. 172 and 173) is usually associated with erythroderma. The patient presents with fever, malaise, myalgias, nausea, vomiting, diarrhea, and confusion. There is a sunburn-type rash that may be subtle and patchy but is usually diffuse and is found on the face, trunk, and extremities. Erythroderma, which desquamates after 1–2 weeks, is more common in Staphylococcus-associated than in Streptococcus-associated TSS. Hypotension develops rapidly—often within hours—after the onset of symptoms. Multiorgan failure occurs. Early renal failure may precede hypotension and distinguishes this syndrome from other septic shock syndromes. There may be no indication of a primary focal infection, although possible cutaneous or mucosal portals of entry for the organism can be ascertained when a careful history is taken. Colonization rather than overt infection of the vagina or a postoperative wound, for example, is typical with staphylococcal TSS, and the mucosal areas appear hyperemic but not infected. Streptococcal TSS is more often associated with skin or soft tissue infection (including necrotizing fasciitis), and patients are more likely to be bacteremic. TSS caused by Clostridium sordellii is associated with childbirth or with skin injection of black-tar heroin. The diagnosis of TSS is defined by the clinical criteria of fever, rash, hypotension, and multiorgan involvement. The mortality rate is 5% for menstruation-associated TSS, 10–15% for nonmenstrual TSS, 30–70% for streptococcal TSS, and up to 90% for obstetric C. sordellii TSS.
![]() Viral Hemorrhagic Fevers Viral hemorrhagic fevers (Chaps. 233 and 234) are zoonotic illnesses caused by viruses that reside in either animal reservoirs or arthropod vectors. These diseases occur worldwide and are restricted to areas where the host species live. They are caused by four major groups of viruses: Arenaviridae (e.g., Lassa fever in Africa), Bunyaviridae (e.g., Rift Valley fever in Africa; hantavirus hemorrhagic fever with renal syndrome in Asia; or Crimean-Congo hemorrhagic fever, which has an extensive geographic distribution), Filoviridae (e.g., Ebola and Marburg virus infections in Africa), and Flaviviridae (e.g., yellow fever in Africa and South America and dengue in Asia, Africa, and the Americas). Lassa fever and Ebola and Marburg virus infections are also transmitted from person to person. The vectors for most viral fevers are found in rural areas; dengue and yellow fever are important exceptions. After a prodrome of fever, myalgias, and malaise, patients develop evidence of vascular damage, petechiae, and local hemorrhage. Shock, multifocal hemorrhaging, and neurologic signs (e.g., seizures or coma) predict a poor prognosis. Dengue (Chap. 233) is the most common arboviral disease worldwide. More than half a million cases of dengue hemorrhagic fever occur each year, with at least 12,000 deaths. Patients have a triad of symptoms: hemorrhagic manifestations, evidence of plasma leakage, and platelet counts of <100,000/μL. Mortality rates are 10–20%. If dengue shock syndrome develops, mortality rates can reach 40%. Supportive care to maintain blood pressure and intravascular volume with careful volume-replacement therapy is key to survival. Ribavirin also may be useful against Arenaviridae and Bunyaviridae.
Viral Hemorrhagic Fevers Viral hemorrhagic fevers (Chaps. 233 and 234) are zoonotic illnesses caused by viruses that reside in either animal reservoirs or arthropod vectors. These diseases occur worldwide and are restricted to areas where the host species live. They are caused by four major groups of viruses: Arenaviridae (e.g., Lassa fever in Africa), Bunyaviridae (e.g., Rift Valley fever in Africa; hantavirus hemorrhagic fever with renal syndrome in Asia; or Crimean-Congo hemorrhagic fever, which has an extensive geographic distribution), Filoviridae (e.g., Ebola and Marburg virus infections in Africa), and Flaviviridae (e.g., yellow fever in Africa and South America and dengue in Asia, Africa, and the Americas). Lassa fever and Ebola and Marburg virus infections are also transmitted from person to person. The vectors for most viral fevers are found in rural areas; dengue and yellow fever are important exceptions. After a prodrome of fever, myalgias, and malaise, patients develop evidence of vascular damage, petechiae, and local hemorrhage. Shock, multifocal hemorrhaging, and neurologic signs (e.g., seizures or coma) predict a poor prognosis. Dengue (Chap. 233) is the most common arboviral disease worldwide. More than half a million cases of dengue hemorrhagic fever occur each year, with at least 12,000 deaths. Patients have a triad of symptoms: hemorrhagic manifestations, evidence of plasma leakage, and platelet counts of <100,000/μL. Mortality rates are 10–20%. If dengue shock syndrome develops, mortality rates can reach 40%. Supportive care to maintain blood pressure and intravascular volume with careful volume-replacement therapy is key to survival. Ribavirin also may be useful against Arenaviridae and Bunyaviridae.
SEPSIS WITH A SOFT TISSUE/MUSCLE PRIMARY FOCUS
See also Chap. 156.
Necrotizing Fasciitis This infection is characterized by extensive necrosis of the subcutaneous tissue and fascia. It may arise at a site of minimal trauma or postoperative incision and may also be associated with recent varicella, childbirth, or muscle strain. The most common causes of necrotizing fasciitis are group A streptococci alone (Chap. 173), the incidence of which has been increasing for the past two decades, and a mixed facultative and anaerobic flora (Chap. 156). Diabetes mellitus, IV drug use, chronic liver or renal disease, and malignancy are associated risk factors. Physical findings are initially minimal compared with the severity of pain and the degree of fever. The examination is often unremarkable except for soft tissue edema and erythema. The infected area is red, hot, shiny, swollen, and exquisitely tender. In untreated infection, the overlying skin develops blue-gray patches after 36 h, and cutaneous bullae and necrosis develop after 3–5 days. Necrotizing fasciitis due to a mixed flora, but not that due to group A streptococci, can be associated with gas production. Without treatment, pain decreases because of thrombosis of the small blood vessels and destruction of the peripheral nerves—an ominous sign. The mortality rate is 15–34% overall, >70% in association with TSS, and nearly 100% without surgical intervention. Necrotizing fasciitis may also be due to Clostridium perfringens (Chap. 179); in this condition, the patient is extremely toxic and the mortality rate is high. Within 48 h, rapid tissue invasion and systemic toxicity associated with hemolysis and death ensue. The distinction between this entity and clostridial myonecrosis is made by muscle biopsy. Necrotizing fasciitis caused by community-acquired MRSA also has been reported.
Clostridial Myonecrosis (See also Chap. 179) Myonecrosis is often associated with trauma or surgery but can develop spontaneously. The incubation period is usually 12–24 h long, and massive necrotizing gangrene develops within hours of onset. Systemic toxicity, shock, and death can occur within 12 h. The patient’s pain and toxic appearance are out of proportion to physical findings. On examination, the patient is febrile, apathetic, tachycardic, and tachypneic and may express a feeling of impending doom. Hypotension and renal failure develop later, and hyperalertness is evident preterminally. The skin over the affected area is bronze-brown, mottled, and edematous. Bullous lesions with serosanguineous drainage and a mousy or sweet odor can develop. Crepitus can occur secondary to gas production in muscle tissue. The mortality rate is >65% for spontaneous myonecrosis, which is often associated with Clostridium septicum or C. tertium and underlying malignancy. The mortality rates associated with trunk and limb infection are 63% and 12%, respectively, and any delay in surgical treatment increases the risk of death.
NEUROLOGIC INFECTIONS WITH OR WITHOUT SEPTIC SHOCK
Bacterial Meningitis (See also Chap. 164) Bacterial meningitis is one of the most common infectious disease emergencies involving the central nervous system. Although hosts with cell-mediated immune deficiency (including transplant recipients, diabetic patients, elderly patients, and cancer patients receiving certain chemotherapeutic agents) are at particular risk for Listeria monocytogenes meningitis, most cases in adults are due to S. pneumoniae (30–60%) and N. meningitidis (10–35%). The classic presentation of fever, meningismus, and altered mental status is seen in only one-half to two-thirds of patients. The elderly can present without fever or meningeal signs. Cerebral dysfunction is evidenced by confusion, delirium, and lethargy that can progress to coma. In some cases, the presentation is fulminant, with sepsis and brain edema; papilledema at presentation is unusual and suggests another diagnosis (e.g., an intracranial lesion). Focal signs, including cranial nerve palsies (IV, VI, VII), can be seen in 10–20% of cases; 50–70% of patients have bacteremia. A poor outcome is associated with coma, hypotension, a pneumococcal etiology, respiratory distress, a CSF glucose level of <0.6 mmol/L (<<0 mg/dL), a CSF protein level of >2.5 g/L, a peripheral white blood cell count of <5000/μL, and a serum sodium level of <135 mmol/L. Rapid initiation of treatment is essential; the odds of an unfavorable outcome may increase by 30% for each hour that treatment is delayed. Mortality also increases linearly with age of the patient.
Suppurative Intracranial Infections (See also Chap. 164) In suppurative intracranial infections, rare intracranial lesions present along with sepsis and hemodynamic instability. Rapid recognition of the toxic patient with central neurologic signs is crucial to improvement of the dismal prognosis of these entities. Subdural empyema arises from the paranasal sinus in 60–70% of cases. Microaerophilic streptococci and staphylococci are the predominant etiologic organisms. The patient is toxic, with fever, headache, and nuchal rigidity. Of all patients, 75% have focal signs and 6–20% die. Despite improved survival rates, 15–44% of patients are left with permanent neurologic deficits. Septic cavernous sinus thrombosis follows a facial or sphenoid sinus infection; 70% of cases are due to staphylococci (including MRSA), and the remainder are due primarily to aerobic or anaerobic streptococci. A unilateral or retroorbital headache progresses to a toxic appearance and fever within days. Three-quarters of patients have unilateral periorbital edema that becomes bilateral and then progresses to ptosis, proptosis, ophthalmoplegia, and papilledema. The mortality rate is as high as 30%. Septic thrombosis of the superior sagittal sinus spreads from the ethmoid or maxillary sinuses and is caused by S. pneumoniae, other streptococci, and staphylococci. The fulminant course is characterized by headache, nausea, vomiting, rapid progression to confusion and coma, nuchal rigidity, and brainstem signs. If the sinus is totally thrombosed, the mortality rate exceeds 80%.
Brain Abscess (See also Chap. 164) Brain abscess often occurs without systemic signs. Almost half of patients are afebrile, and presentations are more consistent with a space-occupying lesion in the brain; 70% of patients have headache and/or altered mental status, 50% have focal neurologic signs, and 25% have papilledema. Abscesses can present as single or multiple lesions resulting from contiguous foci or hematogenous infection, such as endocarditis. The infection progresses over several days from cerebritis to an abscess with a mature capsule. More than half of infections are polymicrobial, with an etiology consisting of aerobic bacteria (primarily streptococcal species) and anaerobes. Abscesses arising hematogenously are especially apt to rupture into the ventricular space, causing a sudden and severe deterioration in clinical status and a high mortality rate. Otherwise, mortality is low but morbidity is high (30–55%). Patients presenting with stroke and a parameningeal infectious focus, such as sinusitis or otitis, may have a brain abscess, and physicians must maintain a high level of suspicion. Prognosis worsens in patients with a fulminant course, delayed diagnosis, abscess rupture into the ventricles, multiple abscesses, or abnormal neurologic status at presentation.
Cerebral Malaria (See also Chap. 248) This entity should be urgently considered if patients who have recently traveled to areas endemic for malaria present with a febrile illness and lethargy or other neurologic signs. Fulminant malaria is caused by Plasmodium falciparum and is associated with temperatures of >40°C (>104°F), hypotension, jaundice, adult respiratory distress syndrome, and bleeding. By definition, any patient with a change in mental status or repeated seizure in the setting of fulminant malaria has cerebral malaria. In adults, this nonspecific febrile illness progresses to coma over several days; occasionally, coma occurs within hours and death within 24 h. Nuchal rigidity and photophobia are rare. On physical examination, symmetric encephalopathy is typical, and upper motor neuron dysfunction with decorticate and decerebrate posturing can be seen in advanced disease. Unrecognized infection results in a 20–30% mortality rate.
Intracranial and Spinal Epidural Abscesses (See also Chap. 456) Spinal and intracranial epidural abscesses (SEAs and ICEAs) can result in permanent neurologic deficits, sepsis, and death. At-risk patients include those with diabetes mellitus; IV drug use; chronic alcohol abuse; recent spinal trauma, surgery, or epidural anesthesia; and other comorbid conditions, such as HIV infection. Fungal epidural abscess and meningitis can follow epidural or paraspinal glucocorticoid infections. In the United States and Canada, where early treatment of otitis and sinusitis is typical, ICEA is rare but the number of cases of SEA is on the rise. In Africa and areas with limited access to health care, SEAs and ICEAs cause significant morbidity and mortality. ICEAs typically present as fever, mental status changes, and neck pain, while SEAs often present as fever, localized spinal tenderness, and back pain. ICEAs are typically polymicrobial, whereas SEAs are most often due to hematogenous seeding, with staphylococci the most common etiologic agent. Early diagnosis and treatment, which may include surgical drainage, minimize rates of mortality and permanent neurologic sequelae. Outcomes are worse for SEA due to MRSA, infection at a higher vertebral-body level, impaired neurologic status on presentation, and dorsal rather than ventral location of the abscess. Elderly patients and persons with renal failure, malignancy, and other comorbidities also have less favorable outcomes.
Other Focal Syndromes with a Fulminant Course Infection at virtually any primary focus (e.g., osteomyelitis, pneumonia, pyelonephritis, or cholangitis) can result in bacteremia and sepsis. Lemierre’s disease—jugular septic thrombophlebitis caused by Fusobacterium necrophorum—is associated with metastatic infectious emboli (primarily to the lung) and sepsis, with mortality rates of >15%. TSS has been associated with focal infections such as septic arthritis, peritonitis, sinusitis, and wound infection. Rapid clinical deterioration and death can be associated with destruction of the primary site of infection, as is seen in endocarditis and in infections of the oropharynx (e.g., Ludwig’s angina or epiglottitis, in which edema suddenly compromises the airway).
Rhinocerebral Mucormycosis (See also Chap. 242) Individuals with diabetes or immunocompromising conditions are at risk for invasive rhinocerebral mucormycosis. Patients present with low-grade fever, dull sinus pain, diplopia, decreased mental status, decreased ocular motion, chemosis, proptosis, dusky or necrotic nasal turbinates, and necrotic hard-palate lesions that respect the midline. Without rapid recognition and intervention, the process continues on an inexorable invasive course, with high mortality rates.
Acute Bacterial Endocarditis (See also Chap. 155) This entity presents with a much more aggressive course than subacute endocarditis. Bacteria such as S. aureus, S. pneumoniae, L. monocytogenes, Haemophilus species, and streptococci of groups A, B, and G attack native valves. Native-valve endocarditis caused by S. aureus (including MRSA strains) is increasing, particularly in health care settings. Mortality rates range from 10% to 40%. The host may have comorbid conditions such as underlying malignancy, diabetes mellitus, IV drug use, or alcoholism. The patient presents with fever, fatigue, and malaise <2 weeks after onset of infection. On physical examination, a changing murmur and congestive heart failure may be noted. Hemorrhagic macules on palms or soles (Janeway lesions) sometimes develop. Petechiae, Roth’s spots, splinter hemorrhages, and splenomegaly are unusual. Rapid valvular destruction, particularly of the aortic valve, results in pulmonary edema and hypotension. Myocardial abscesses can form, eroding through the septum or into the conduction system and causing life-threatening arrhythmias or high-degree conduction block. Large friable vegetations can result in major arterial emboli, metastatic infection, or tissue infarction. Older patients with S. aureus endocarditis are especially likely to present with nonspecific symptoms—a circumstance that delays diagnosis and worsens prognosis. Rapid intervention is crucial for a successful outcome.
Inhalational Anthrax (See also Chap. 261e) Inhalational anthrax, the most severe form of disease caused by B. anthracis, had not been reported in the United States for more than 25 years until the use of this organism as an agent of bioterrorism in 2001. Patients presented with malaise, fever, cough, nausea, drenching sweats, shortness of breath, and headache. Rhinorrhea was unusual. All patients had abnormal chest roentgenograms at presentation. Pulmonary infiltrates, mediastinal widening, and pleural effusions were the most common findings. Hemorrhagic meningitis was seen in 38% of these patients. Survival was more likely when antibiotics were given during the prodromal period and when multidrug regimens were used. In the absence of urgent intervention with antimicrobial agents and supportive care, inhalational anthrax progresses rapidly to hypotension, cyanosis, and death.
Avian and Swine Influenza (See also Chap. 224) Human cases of avian influenza have occurred primarily in Southeast Asia, particularly Vietnam (H5N1) and China (H7N9). Avian influenza should be considered in patients with severe respiratory tract illness, particularly if they have been exposed to poultry. Patients present with high fever, an influenza-like illness, and lower respiratory tract symptoms; this illness can progress rapidly to bilateral pneumonia, acute respiratory distress syndrome, multiorgan failure, and death. Early antiviral treatment with neuraminidase inhibitors should be initiated along with aggressive supportive measures. Unlike avian influenza, for which human-to-human transmission has been rare so far and has not been sustained, a novel swine-associated influenza A/H1N1 virus has spread rapidly throughout the world. Patients most at risk of severe disease are children <5 years of age, elderly persons, patients with underlying chronic conditions, and pregnant women. Obesity also has been identified as a risk factor for severe illness.
Hantavirus Pulmonary Syndrome (See also Chap. 233) Hantavirus pulmonary syndrome has been documented in the United States (primarily the southwestern states), Canada, and South America. Most cases occur in rural areas and are associated with exposure to rodents. Patients present with a nonspecific viral prodrome of fever, malaise, myalgias, nausea, vomiting, and dizziness that may progress to pulmonary edema and respiratory failure. Hantavirus pulmonary syndrome causes myocardial depression and increased pulmonary vascular permeability; therefore, careful fluid resuscitation and use of pressor agents are crucial. Aggressive cardiopulmonary support during the first few hours of illness can be life-saving. The early onset of thrombocytopenia may help distinguish this syndrome from other febrile illnesses in an appropriate epidemiologic setting.
CONCLUSION
Acutely ill febrile patients with the syndromes discussed in this chapter require close observation, aggressive supportive measures, and—in most cases—admission to intensive care units. The most important task of the physician is to distinguish these patients from other infected febrile patients whose illness will not progress to fulminant disease. The alert physician must recognize the acute infectious disease emergency and then proceed with appropriate urgency.
148 |
Immunization Principles and Vaccine Use |
Few medical interventions of the past century can rival the effect that immunization has had on longevity, economic savings, and quality of life. Seventeen diseases are now preventable through vaccines routinely administered to children and adults in the United States (Table 148-1), and most vaccine-preventable diseases of childhood are at historically low levels (Table 148-2). Health care providers deliver the vast majority of vaccines in the United States in the course of providing routine health services and therefore play an integral role in the nation’s public health system.
|
DISEASES PREVENTABLE WITH VACCINES ROUTINELY ADMINISTERED IN THE UNITED STATES TO CHILDREN AND/OR ADULTS |
|
DECLINE IN VACCINE-PREVENTABLE DISEASES IN THE UNITED STATES FOLLOWING WIDESPREAD IMPLEMENTATION OF NATIONAL VACCINE RECOMMENDATIONS |
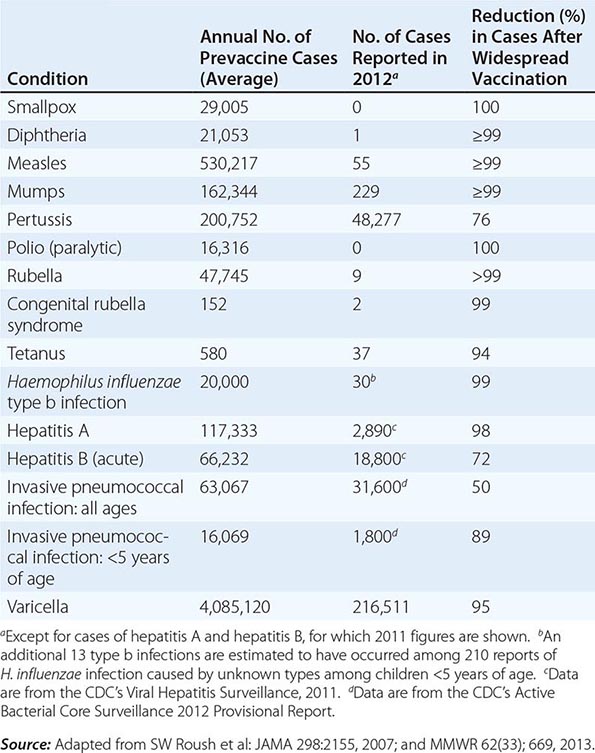
VACCINE IMPACT
Direct and Indirect Effects Immunizations against specific infectious diseases protect individuals against infection and thereby prevent symptomatic illnesses. Specific vaccines may blunt the severity of clinical illness (e.g., rotavirus vaccines and severe gastroenteritis) or reduce complications (e.g., zoster vaccines and postherpetic neuralgia). Some immunizations also reduce transmission of infectious disease agents from immunized people to others, thereby reducing the impact of infection spread. This indirect impact is known as herd immunity. The level of immunization in a population that is required to achieve indirect protection of unimmunized people varies substantially with the specific vaccine.
Since childhood vaccines have become widely available in the United States, major declines in rates of vaccine-preventable diseases among both children and adults have become evident (Table 148-2). For example, vaccination of children <5 years of age against seven types of Streptococcus pneumoniae led to a >90% overall reduction in invasive disease caused by those types. A series of childhood vaccines targeting 13 vaccine-preventable diseases in a single birth cohort leads to prevention of 42,000 premature deaths and 20 million illnesses and saves nearly $70 billion (U.S.).
Control, Elimination, and Eradication of Vaccine-Preventable Diseases Immunization programs are associated with the goals of controlling, eliminating, or eradicating a disease. Control of a vaccine-preventable disease reduces poor illness outcomes and often limits the disruptive impacts associated with outbreaks of disease in communities, schools, and institutions. Control programs can also reduce absences from work for ill persons and for parents caring for sick children, decrease absences from school, and limit health care utilization associated with treatment visits.
Elimination of a disease is a more demanding goal than control, usually requiring the reduction to zero of cases in a defined geographic area but sometimes defined as reduction in the indigenous sustained transmission of an infection in a geographic area. As of 2013, the United States had eliminated indigenous transmission of measles, rubella, poliomyelitis, and diphtheria. Importation of pathogens from other parts of the world continues to be important, and public health efforts are intended to react promptly to such cases and to limit forward spread of the infectious agent.
![]() Eradication of a disease is achieved when its elimination can be sustained without ongoing interventions. The only vaccine-preventable disease of humans that has been globally eradicated thus far is smallpox. Although smallpox vaccine is no longer given routinely, the disease has not reemerged naturally because all chains of human transmission were interrupted through earlier vaccination efforts and humans were the only natural reservoir of the virus. Currently, a major health initiative is targeting the global eradication of polio. Sustained transmission of polio has been eliminated from most nations but has never been interrupted in three countries—Afghanistan, Nigeria, and Pakistan—while recent outbreaks in Syria and the Horn of Africa underscore that other countries remain at risk for importation until these reservoirs have been addressed. Detection of a case of disease that has been targeted for eradication or elimination is considered a sentinel event that could permit the infectious agent to become reestablished in the community or region. Therefore, such episodes must be promptly reported to public health authorities.
Eradication of a disease is achieved when its elimination can be sustained without ongoing interventions. The only vaccine-preventable disease of humans that has been globally eradicated thus far is smallpox. Although smallpox vaccine is no longer given routinely, the disease has not reemerged naturally because all chains of human transmission were interrupted through earlier vaccination efforts and humans were the only natural reservoir of the virus. Currently, a major health initiative is targeting the global eradication of polio. Sustained transmission of polio has been eliminated from most nations but has never been interrupted in three countries—Afghanistan, Nigeria, and Pakistan—while recent outbreaks in Syria and the Horn of Africa underscore that other countries remain at risk for importation until these reservoirs have been addressed. Detection of a case of disease that has been targeted for eradication or elimination is considered a sentinel event that could permit the infectious agent to become reestablished in the community or region. Therefore, such episodes must be promptly reported to public health authorities.
Outbreak Detection and Control Clusters of cases of a vaccine-preventable disease detected in an institution, a medical practice, or a community may signal important changes in the pathogen, vaccine, or environment. Several factors can give rise to increases in vaccine-preventable disease, including (1) low rates of immunization that result in an accumulation of susceptible people (e.g., measles resurgence among vaccination abstainers); (2) changes in the infectious agent that permit it to escape vaccine-induced protection (e.g., non-vaccine-type pneumococci); (3) waning of vaccine-induced immunity (e.g., pertussis among adolescents and adults vaccinated in early childhood); and (4) point-source introductions of large inocula (e.g., food-borne exposure to hepatitis A virus). Reporting episodes of outbreak-prone diseases to public health authorities can facilitate recognition of clusters that require further interventions.
PUBLIC HEALTH REPORTING Recognition of suspected cases of diseases targeted for elimination or eradication—along with other diseases that require urgent public health interventions, such as contact tracing, administration of chemo- or immunoprophylaxis, or epidemiologic investigation for common-source exposure—is typically associated with special reporting requirements. Many diseases against which vaccines are routinely used, including measles, pertussis, Haemophilus influenzae type b invasive disease, and varicella, are nationally notifiable. Clinicians and laboratory staff have a responsibility to report some vaccine-preventable disease occurrences to local or state public health authorities according to specific case-definition criteria. All providers should be aware of state or city disease-reporting requirements and the best ways to contact public health authorities. A prompt response to vaccine-preventable disease outbreaks can greatly enhance the effectiveness of control measures.
![]() GLOBAL CONSIDERATIONS Several international health initiatives currently focus on reducing vaccine-preventable diseases in regions throughout the world. These efforts include improving access to new and underutilized vaccines, such as pneumococcal conjugate, rotavirus, human papillomavirus (HPV), and meningococcal A conjugate vaccines. The American Red Cross, the World Health Organization (WHO), the United Nations Foundation, the United Nations Children’s Fund (UNICEF), and the Centers for Disease Control and Prevention (CDC) are partners in the Measles & Rubella Initiative, which targets reduction of worldwide measles deaths by 95% from 2000 to 2015. During 2000–2011, global measles mortality rates declined by 71%—i.e., from an estimated 548,000 deaths in 2000 to 158,000 deaths in 2011. Rotary International, UNICEF, the CDC, and the WHO are leading partners in the global eradication of polio, an endeavor that reduced the annual number of paralytic polio cases from 350,000 in 1988 to <250 in 2012. The GAVI Alliance and the Bill and Melinda Gates Foundation have brought substantial momentum to global efforts to reduce vaccine-preventable diseases, expanding on earlier efforts by the WHO, UNICEF, and governments in developed and developing countries.
GLOBAL CONSIDERATIONS Several international health initiatives currently focus on reducing vaccine-preventable diseases in regions throughout the world. These efforts include improving access to new and underutilized vaccines, such as pneumococcal conjugate, rotavirus, human papillomavirus (HPV), and meningococcal A conjugate vaccines. The American Red Cross, the World Health Organization (WHO), the United Nations Foundation, the United Nations Children’s Fund (UNICEF), and the Centers for Disease Control and Prevention (CDC) are partners in the Measles & Rubella Initiative, which targets reduction of worldwide measles deaths by 95% from 2000 to 2015. During 2000–2011, global measles mortality rates declined by 71%—i.e., from an estimated 548,000 deaths in 2000 to 158,000 deaths in 2011. Rotary International, UNICEF, the CDC, and the WHO are leading partners in the global eradication of polio, an endeavor that reduced the annual number of paralytic polio cases from 350,000 in 1988 to <250 in 2012. The GAVI Alliance and the Bill and Melinda Gates Foundation have brought substantial momentum to global efforts to reduce vaccine-preventable diseases, expanding on earlier efforts by the WHO, UNICEF, and governments in developed and developing countries.
Enhancing Immunization in Adults Although immunization has become a centerpiece of routine pediatric medical visits, it has not been as well integrated into routine health care visits for adults. This chapter focuses on immunization principles and vaccine use in adults. Accumulating evidence suggests that immunization coverage can be increased through efforts directed at consumer-, provider-, institution-, and system-level factors. The literature suggests that the application of multiple strategies is more effective at raising coverage rates than is the use of any single strategy.
RECOMMENDATIONS FOR ADULT IMMUNIZATIONS The CDC’s Advisory Committee on Immunization Practices (ACIP) is the main source of recommendations for administration of vaccines approved by the U.S. Food and Drug Administration (FDA) for use in children and adults in the U.S. civilian population. The ACIP is a federal advisory committee that consists of 15 voting members (experts in fields associated with immunization) appointed by the Secretary of the U.S. Department of Health and Human Services; 8 ex officio members representing federal agencies; and 26 nonvoting representatives of various liaison organizations, including major medical societies and managed-care organizations. The ACIP recommendations are available at www.cdc.gov/vaccines/hcp/acip-recs/. These recommendations are harmonized to the greatest extent possible with vaccine recommendations made by other organizations, including the American College of Obstetricians and Gynecologists, the American Academy of Family Physicians, and the American College of Physicians.
ADULT IMMUNIZATION SCHEDULES Immunization schedules for adults in the United States are updated annually and can be found online (www.cdc.gov/vaccines/schedules/hcp/adult.html). In January, the schedules are published in American Family Physician, Annals of Internal Medicine, and Morbidity and Mortality Weekly Report (www.cdc.gov/mmwr). The adult immunization schedules for 2013 are summarized in Fig. 148-1. Additional information and specifications are contained in the footnotes to these schedules. In the time between annual publications, additions and changes to schedules are published as Notices to Readers in Morbidity and Mortality Weekly Report.
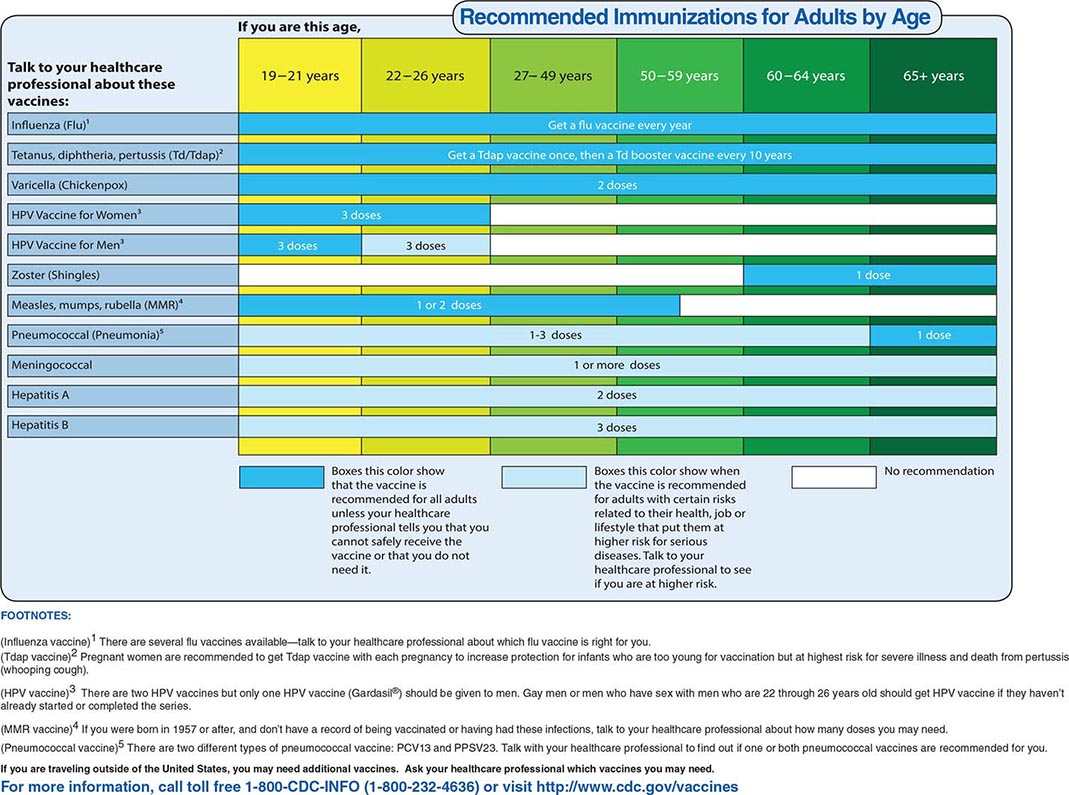
FIGURE 148-1 Recommended adult immunization schedules, United States, 2013. For complete statements by the Advisory Committee on Immunization Practices (ACIP), visit www.cdc.gov/vaccines/hcp/acip-recs/.
IMMUNIZATION PRACTICE STANDARDS
Administering immunizations to adults involves a number of processes, such as deciding whom to vaccinate, assessing vaccine contraindications and precautions, providing vaccine information statements (VISs), ensuring appropriate storage and handling of vaccines, administering vaccines, and maintaining vaccine records. In addition, provider reporting of adverse events that follow vaccination is an essential component of the vaccine safety monitoring system.
Deciding Whom to Vaccinate Every effort should be made to ensure that adults receive all indicated vaccines as expeditiously as possible. When adults present for care, their immunization history should be assessed and recorded, and this information should be used to identify needed vaccinations according to the most current version of the adult immunization schedule. Decision-support tools incorporated into electronic health records can provide prompts for needed vaccinations. Standing orders, which are often used for routinely indicated vaccines (e.g., influenza and pneumococcal vaccines), permit a nurse or another approved licensed practitioner to administer vaccines without a specific physician order, thus lowering barriers to adult immunization.
Assessing Contraindications and Precautions Before vaccination, all patients should be screened for contraindications and precautions. A contraindication is a condition that increases the risk of a serious adverse reaction to vaccination. A vaccine should not be administered when a contraindication is documented. For example, a history of an anaphylactic reaction to a dose of vaccine or to a vaccine component is a contraindication for further doses. A precaution is a condition that may increase the risk of an adverse event or that may compromise the ability of the vaccine to evoke immunity (e.g., administering measles vaccine to a person who has recently received a blood transfusion and may consequently have transient passive immunity to measles virus). Normally, a vaccine is not administered when a precaution is noted. However, situations may arise when the benefits of vaccination outweigh the estimated risk of an adverse event, and the provider may decide to vaccinate the patient despite the precaution.
In some cases, contraindications and precautions are temporary and may lead to mere deferral of vaccination until a later time. For example, moderate or severe acute illness with or without fever is generally considered a transient precaution to vaccination and results in postponement of vaccine administration until the acute phase has resolved; thus the superimposition of adverse effects of vaccination on the underlying illness and the mistaken attribution of a manifestation of the underlying illness to the vaccine are avoided. Contraindications and precautions to vaccines licensed in the United States for use in civilian adults are summarized in Table 148-3. It is important to recognize conditions that are not contraindications in order not to miss opportunities for vaccination. For example, in most cases, mild acute illness (with or without fever), a history of a mild to moderate local reaction to a previous dose of the vaccine, and breast-feeding are not contraindications to vaccination.
|
CONTRAINDICATIONS AND PRECAUTIONS FOR COMMONLY USED VACCINES IN ADULTS |
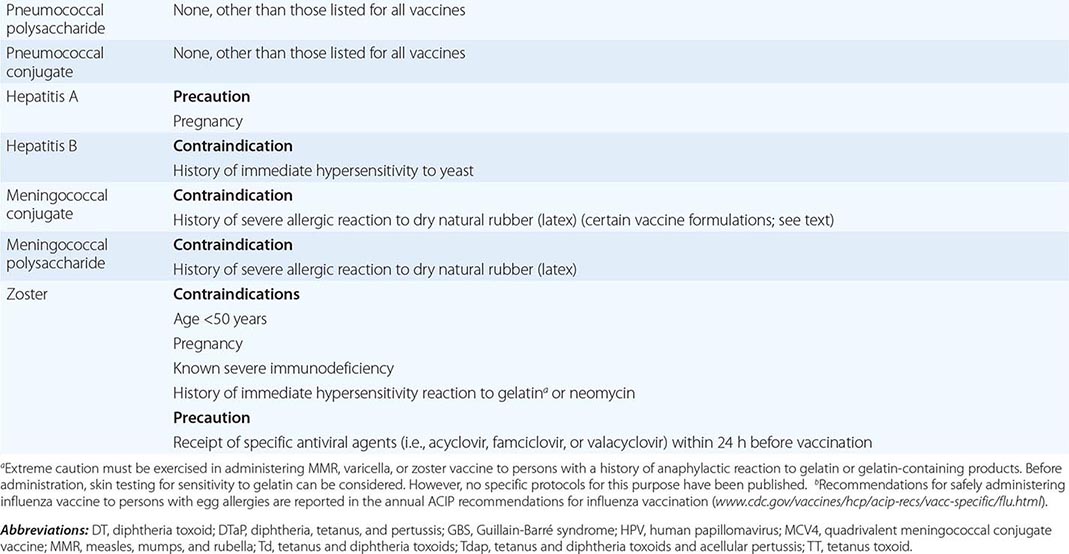
HISTORY OF IMMEDIATE HYPERSENSITIVITY TO A VACCINE COMPONENT A severe allergic reaction (e.g., anaphylaxis) to a previous dose of a vaccine or to one of its components is a contraindication to vaccination. While most vaccines have many components, substances to which individuals are most likely to have had a severe allergic reaction include egg protein, gelatin, and yeast. In addition, although natural rubber (latex) is not a vaccine component, some vaccines are supplied in vials or syringes that contain natural rubber latex. These vaccines can be identified by the product insert and should not be administered to persons who report a severe (anaphylactic) allergy to latex unless the benefit of vaccination clearly outweighs the risk for a potential allergic reaction. The much more common local or contact hypersensitivity to latex, such as to medical gloves (which contain synthetic latex that is not linked to allergic reactions), is not a contraindication to administration of a vaccine supplied in a vial or syringe that contains natural rubber latex. Vaccines routinely indicated for adults that, as of December 2012, were sometimes supplied in a vial or syringe containing natural rubber include Havrix hepatitis A vaccine (syringe); Vaqta hepatitis A vaccine (vial and syringe); Engerix-B hepatitis B vaccine (syringe); Recombivax HB hepatitis B vaccine (vial); Cervarix HPV vaccine (syringe); Fluarix, Fluvirin, Agriflu, and Flucelvax influenza vaccines (syringe); Adacel and Boostrix Tdap (tetanus and diphtheria toxoids and acellular pertussis) vaccines (syringe); Td (tetanus and diphtheria toxoids) vaccines (syringe); Twinrix hepatitis A and B vaccine (syringe); and Menomune meningococcal polysaccharide vaccine (vial).
PREGNANCY Live-virus vaccines are contraindicated during pregnancy because of the theoretical risk that vaccine virus replication will cause congenital infection or have other adverse effects on the fetus. Most live-virus vaccines, including varicella vaccine, are not secreted in breast milk; therefore, breast-feeding is not a contraindication for live-virus or other vaccines. Pregnancy is not a contraindication to administration of inactivated vaccines, but most are avoided during pregnancy because relevant safety data are limited. Two inactivated vaccines, Tdap vaccine and inactivated influenza vaccine, are routinely recommended for pregnant women in the United States. Tdap vaccine is recommended during each pregnancy, regardless of prior vaccination status, in order to prevent pertussis in neonates. Annual influenza vaccination is recommended for all persons 6 months of age and older, regardless of pregnancy status. Some other vaccines, such as meningococcal vaccines, may be given to pregnant women in certain circumstances.
IMMUNOSUPPRESSION Live-virus vaccines elicit an immune response due to replication of the attenuated (weakened) vaccine virus that is contained by the recipient’s immune system. In persons with compromised immune function, enhanced replication of vaccine viruses is possible and could lead to disseminated infection with the vaccine virus. For this reason, live-virus vaccines are contraindicated for persons with severe immunosuppression, the definition of which may vary with the vaccine. Severe immunosuppression may be caused by many disease conditions, including HIV infection and hematologic or generalized malignancy. In some of these conditions, all affected persons are severely immunocompromised. In others (e.g., HIV infection), the degree to which the immune system is compromised depends on the severity of the condition, which in turn depends on the stage of disease or treatment. For example, measles-mumps-rubella (MMR) vaccine may be given to HIV-infected persons who are not severely immunocompromised. Severe immunosuppression may also be due to therapy with immunosuppressive agents, including high-dose glucocorticoids. In this situation, the dose, duration, and route of administration may influence the degree of immunosuppression.
VACCINE INFORMATION STATEMENTS
A VIS is a one-page (two-sided) information sheet produced by the CDC that informs vaccine recipients (or their parents or legal representatives) about the benefits and risks of a vaccine. VISs are mandated by the National Childhood Vaccine Injury Act (NCVIA) of 1986 and—whether the vaccine recipient is a child or an adult—must be provided for any vaccine covered by the Vaccine Injury Compensation Program. As of July 2011, vaccines that are covered by the NCVIA and that are licensed for use in adults include Td, Tdap, hepatitis A, hepatitis B, HPV, trivalent inactivated influenza, trivalent live intranasal influenza, MMR, 13-valent pneumococcal conjugate, meningococcal, polio, and varicella vaccines. When combination vaccines for which no separate VIS exists are given (e.g., hepatitis A and B combination vaccine), all relevant VISs should be provided. VISs also exist for some vaccines not covered by the NCVIA, such as pneumococcal polysaccharide, Japanese encephalitis, rabies, zoster, typhoid, and yellow fever vaccines. The use of these VISs is encouraged but is not mandated.
All current VISs are available on the Internet at two websites: the CDC’s Vaccines & Immunizations site (www.cdc.gov/vaccines/hcp/vis/) and the Immunization Action Coalition’s site (www.immunize.org/vis/). (The latter site also includes translations of the VISs.) VISs from these sites can be downloaded and printed.
STORAGE AND HANDLING
Injectable vaccines are packaged in multidose vials, single-dose vials, or manufacturer-filled single-dose syringes. The live attenuated nasal-spray influenza vaccine is packaged in single-dose sprayers. Oral typhoid vaccine is packaged in capsules. Some vaccines, such as MMR, varicella, zoster, and meningococcal polysaccharide vaccines, come as lyophilized (freeze-dried) powders that must be reconstituted (i.e., mixed with a liquid diluent) before use. The lyophilized powder and the diluent come in separate vials. Diluents are not interchangeable but rather are specifically formulated for each type of vaccine; only the specific diluent provided by the manufacturer for each type of vaccine should be used. Once lyophilized vaccines have been reconstituted, their shelf-life is limited and they must be stored under appropriate temperature and light conditions. For example, varicella and zoster vaccines must be protected from light and administered within 30 min of reconstitution; MMR vaccine likewise must be protected from light but can be used up to 8 h after reconstitution. Single-dose vials of meningococcal polysaccharide vaccine must be used within 30 min of reconstitution, while multidose vials must be used within 35 days.
Vaccines are stored either at refrigerator temperature (2–8°C) or at freezer temperature (–15°C or colder). In general, inactivated vaccines (e.g., inactivated influenza, pneumococcal polysaccharide, and meningococcal conjugate vaccines) are stored at refrigerator temperature, while vials of lyophilized-powder live-virus vaccines (e.g., varicella, zoster, and MMR vaccines) are stored at freezer temperature. Diluents for lyophilized vaccines may be stored at refrigerator or room temperature. Live attenuated influenza vaccine—a live-virus liquid formulation administered by nasal spray—is stored at refrigerator temperature.
Vaccine storage and handling errors can result in the loss of vaccines worth millions of dollars, and administration of improperly stored vaccines may elicit inadequate immune responses in patients. To improve the standard of vaccine storage and handling practices, the CDC has published detailed guidance (available at www.cdc.gov/vaccines/recs/storage/toolkit/storage-handling-toolkit.pdf). For vaccine storage, the CDC recommends stand-alone units—i.e., self-contained units that either refrigerate or freeze but do not do both—as these units maintain the required temperatures better than combination refrigerator/freezer units. Dormitory-style combined refrigerator/freezer units should never be used for vaccine storage.
The temperature of refrigerators and freezers used for vaccine storage must be monitored and the temperature recorded at least twice each workday. Ideally, continuous thermometers that measure and record temperature all day and all night are used, and minimum and maximum temperatures are read and documented each workday. The CDC recommends the use of calibrated digital thermometers with a probe in a glycol-filled bottle; more detailed information on specifications of storage units and temperature-monitoring devices is provided at the link given above.
ADMINISTRATION OF VACCINES
Most parenteral vaccines recommended for routine administration to adults in the United States are given by either the IM or the SC route; one influenza vaccine formulation approved for use in adults 18–64 years of age is given intradermally. Live-virus vaccines such as varicella, zoster, and MMR are given SC. Most inactivated vaccines are given IM, except for meningococcal polysaccharide vaccine, which is given SC. The 23-valent pneumococcal polysaccharide vaccine may be given either IM or SC, but IM administration is preferred because it is associated with a lower risk of injection-site reactions.
Vaccines given to adults by the SC route are administered with a 5/8-inch needle into the upper outer-triceps area. Vaccines administered to adults by the IM route are injected into the deltoid muscle (Fig. 148-2) with a needle whose length should be selected on the basis of the recipient’s sex and weight to ensure adequate penetration into the muscle. Current guidelines indicate that, for men and women weighing <152 lbs (<70 kg), a 1-inch needle is sufficient; for women weighing 152–200 lbs (70–90 kg) and men weighing 152–260 lbs (70–118 kg), a 1- to 1.5-inch needle is needed; and for women weighing >200 lbs (>90 kg) and men weighing >260 lbs (>118 kg), a 1.5-inch needle is required. Additional illustrations of vaccine injection locations and techniques may be found at www.immunize.org/catg.d/p2020a.pdf.
FIGURE 148-2 Technique for IM administration of vaccine. (Photo credit: James Gathany, Centers for Disease Control and Prevention; accessible at Public Health Image Library, www.cdc.gov. PHIL ID#9420.)
Aspiration, the process of pulling back on the plunger of the syringe after skin penetration but prior to injection, is not necessary because no large blood vessels are present at the recommended vaccine injection sites.
Multiple vaccines can be administered at the same visit; indeed, administration of all needed vaccines at one visit is encouraged. Studies have shown that vaccines are as effective when administered simultaneously as they are individually, and simultaneous administration of multiple vaccines is not associated with an increased risk of adverse effects. If more than one vaccine must be administered in the same limb, the injection sites should be separated by 1–2 inches so that any local reactions can be differentiated. If a vaccine and an immune globulin preparation are administered simultaneously (e.g., Td vaccine and tetanus immune globulin), a separate anatomic site should be used for each injection.
For certain vaccines (e.g., HPV vaccine and hepatitis B vaccine), multiple doses are required for an adequate and persistent antibody response. The recommended vaccination schedule specifies the interval between doses. Many adults who receive the first dose in a multiple-dose vaccine series do not complete the series or do not receive subsequent doses within the recommended interval. For example, at least one-third of adults who receive the first dose of hepatitis B vaccine in the three-dose series do not complete the series. In these circumstances, vaccine efficacy and/or the duration of protection may be compromised. Providers should implement recall systems that will prompt patients to return for subsequent doses in a vaccination series at the appropriate intervals. With the exception of oral typhoid vaccination, an interruption in the schedule does not require restarting of the entire series or the addition of extra doses.
Syncope may follow vaccination, especially in adolescents and young adults. Serious injuries, including skull fracture and cerebral hemorrhage, have occurred. Adolescents and adults should be seated or lying down during vaccination. The majority of reported syncope episodes after vaccination occur within 15 min. The ACIP recommends that vaccine providers strongly consider observing patients, particularly adolescents, with patients seated or lying down for 15 min after vaccination. If syncope develops, patients should be observed until the symptoms resolve.
Anaphylaxis is a rare complication of vaccination. All facilities providing immunizations should have an emergency kit containing aqueous epinephrine for administration in the event of a systemic anaphylactic reaction.
MAINTENANCE OF VACCINE RECORDS
All vaccines administered should be fully documented in the patient’s permanent medical record. Documentation should include the date of administration, the name or common abbreviation of the vaccine, the vaccine lot number and manufacturer, the administration site, the VIS edition, the date the VIS was provided, and the name, address, and title of the person who administered the vaccine. Increasing use of two-dimensional bar codes on vaccine vials and syringes that can be scanned for data entry into compatible electronic medical records and immunization information systems may facilitate more complete and accurate recording of required information.
VACCINE SAFETY MONITORING AND ADVERSE EVENT REPORTING
Prelicensure Evaluations of Vaccine Safety Before vaccines are licensed by the FDA, they are evaluated in clinical trials with volunteers. These trials are conducted in three progressive phases. Phase 1 trials are small, usually involving fewer than 100 volunteers. Their purposes are to provide a basic evaluation of safety and to identify common adverse events. Phase 2 trials, which are larger and may involve several hundred participants, collect additional information on safety and are usually designed to evaluate immunogenicity as well. Data gained from phase 2 trials can be used to determine the composition of the vaccine, the number of doses required, and a profile of common adverse events. Vaccines that appear promising are evaluated in phase 3 trials, which typically involve several hundred to several thousand volunteers and are generally designed to demonstrate vaccine efficacy and provide additional information on vaccine safety.
Postlicensure Monitoring of Vaccine Safety After licensure, a vaccine’s safety is assessed by several mechanisms. The NCVIA of 1986 requires health care providers to report certain adverse events that follow vaccination. As a mechanism for that reporting, the Vaccine Adverse Event Reporting System (VAERS) was established in 1990 and is jointly managed by the CDC and the FDA. This safety surveillance system collects reports of adverse events associated with vaccines currently licensed in the United States. Adverse events are defined as untoward events that occur after immunization and that might be caused by the vaccine product or vaccination process. While the VAERS was established in response to the NCVIA, any adverse event following vaccination—whether in a child or an adult, and whether or not it is believed to have actually been caused by vaccination—may be reported through the VAERS. The adverse events that health care providers are required to report are listed in the reportable-events table on the VAERS website at vaers.hhs.gov/reportable.htm. Approximately 30,000 VAERS reports are filed annually, with ~13% reporting serious events resulting in hospitalization, life-threatening illness, disability, or death.
Anyone can file a VAERS report, including health care providers, manufacturers, and vaccine recipients or their parents or guardians. VAERS reports may be submitted online (vaers.hhs.gov/esub/step1) or by completing a paper form requested by email (info@vaers.org), phone (800-822-7967), or fax (877-721-0366). The VAERS form asks for the following information: the type of vaccine received; the timing of vaccination; the time of onset of the adverse event; and the recipient’s current illnesses or medications, history of adverse events following vaccination, and demographic characteristics (e.g., age and sex). This information is entered into a database. The individual who reported the adverse event then receives a confirmation letter by mail with a VAERS identification number that can be used if additional information is submitted later. In selected cases of serious adverse reaction, the patient’s recovery status may be followed up at 60 days and 1 year after vaccination. The FDA and the CDC have access to VAERS data and use this information to monitor vaccine safety and conduct research studies. VAERS data (minus personal information) are also available to the public.
While the VAERS provides useful information on vaccine safety, this passive reporting system has important limitations. One is that events following vaccination are merely reported; the system cannot assess whether a given type of event occurs more often than expected after vaccination. A second is that event reporting is incomplete and is biased toward events that are believed to be more likely to be due to vaccination and that occur relatively soon after vaccination. To obtain more systematic information on adverse events occurring in both vaccinated and unvaccinated persons, the Vaccine Safety Datalink project was initiated in 1991. Directed by the CDC, this project includes nine managed-care organizations in the United States; member databases include information on immunizations, medical conditions, demographics, laboratory results, and medication prescriptions. The Department of Defense oversees a similar system monitoring the safety of immunizations among active-duty military personnel. In addition, postlicensure evaluations of vaccine safety may be conducted by the vaccine manufacturer. In fact, such evaluations are often required by the FDA as a condition of vaccine licensure.
CONSUMER ACCESS TO AND DEMAND FOR IMMUNIZATION
By removing barriers to the consumer or patient, providers and health care institutions can improve vaccine use. Financial barriers have traditionally been important constraints, particularly among uninsured adults. Even for insured adults, out-of-pocket costs associated with newer, more expensive adult vaccines (e.g., zoster vaccine) are an obstacle to be overcome. After influenza vaccine was included by Medicare for all beneficiaries in 1993, coverage among persons ≥65 years of age doubled (from ~30% in 1989 to >60% in 1997). Other strategies that enhance patients’ access to vaccination include extended office hours (e.g., evening and weekend hours) and scheduled vaccination-only clinics where waiting times are reduced. Provision of vaccines outside the “medical home” (e.g., through occupational clinics, universities, pharmacies, and retail settings) can expand access for adults who do not make medical visits frequently. Increasing proportions of adults are being vaccinated in these settings.
Health promotion efforts aimed at increasing the demand for immunization are common. Direct-to-consumer advertising by pharmaceutical companies has been used for some newer adolescent and adult vaccines. Efforts to raise consumer demand for vaccines have not increased immunization rates unless implemented in conjunction with other strategies that target strengthening of provider practices or reduction of consumer barriers. Attitudes and beliefs related to vaccination can be considerable impediments to consumer demand. Many adults view vaccines as important for children but are less familiar with vaccinations targeting disease prevention in adults. Several vaccines are recommended for adults with certain medical risk factors, but self-identification as a high-risk individual is relatively rare. Communication research suggests that many adults with chronic diseases may be more motivated to receive a vaccine by a desire to protect their family members rather than to reduce their own risk. Some vaccines are explicitly recommended for persons at relatively low risk of serious complications, with the goal of reducing the risk of transmission to higher-risk contacts. For example, for protection of newborns, vaccinations against influenza and pertussis are recommended for pregnant women and for others who will be around the infant.
STRATEGIES FOR PROVIDERS AND HEALTH CARE FACILITIES
Recommendation from the Provider Health care providers can have great influence on patients with regard to immunization. A recommendation from a doctor or nurse carries more weight than do recommendations from professional societies or endorsements by celebrities. Providers should be well informed about vaccine risks and benefits so that they can address patients’ common concerns. The CDC, the American College of Physicians, and the American Academy of Family Physicians review and update the schedule for adult immunization on an annual basis and also have developed educational materials to facilitate provider–patient discussions about vaccination (www.cdc.gov/vaccines/hcp.htm).
System Supports Medical offices can incorporate a variety of methods to ensure that providers consistently offer specific immunizations to patients with indications for specific vaccines. Decision-support tools have been incorporated into some electronic health records to alert the provider when specific vaccines are indicated. Manual or automated reminders and standing orders have been discussed (see “Deciding Whom to Vaccinate,” above) and have consistently improved vaccination coverage in both office and hospital settings. Most clinicians’ estimates of their own performance diverge from objective measurements of their patients’ immunization coverage; quantitative assessment and feedback have been shown in pediatric and adolescent practices to increase immunization performance significantly. Some health plans have instituted incentives for providers with high rates of immunization coverage. Specialty providers, including obstetrician–gynecologists, may be the only providers serving some high-risk patients with indications for selected vaccines (e.g., Tdap, influenza, or pneumococcal polysaccharide vaccine).
Immunization Requirements Vaccination against selected communicable diseases is required for attendance at many universities and colleges as well as for service in the U.S. military or in some occupational settings (e.g., child care, laboratory, veterinary, and health care). Immunizations are recommended and sometimes required for travel to certain countries (Chap. 149).
Vaccination of Health Care Staff A particular area of focus for medical settings is vaccination of health care workers, including those with and without direct patient-care responsibilities. The Joint Commission (which accredits health care organizations), the CDC’s Healthcare Infection Control Practices Advisory Committee, and the ACIP all recommend influenza vaccination of all health care personnel; recommendations also focus on requiring documentation of declination for providers who do not accept annual influenza vaccination. As part of their participation in the Centers for Medicare and Medicaid Services’ Hospital Inpatient Quality Reporting program, acute-care hospitals are required to report the proportion of their health care personnel who have received seasonal influenza vaccine. Some institutions and jurisdictions have added mandates on influenza vaccination of health care workers and have expanded on earlier requirements related to vaccination or proof of immunity for hepatitis B, measles, mumps, rubella, and varicella.
VACCINATION IN NONMEDICAL SETTINGS
Receipt of vaccination in medical offices is most frequent among young children and adults ≥65 years of age. People in these age groups make more office visits and are more likely to receive care in a consistent “medical home” than are older children, adolescents, and nonelderly adults. Vaccination outside the medical home can expand access to those whose health care visits are limited and reduce the burden on busy clinical practices. In some locations, financial constraints related to inventory and storage requirements have led providers to stock few or no vaccines. Outside private office and hospital settings, vaccination may also occur at health department venues, workplaces, retail sites (including pharmacies and supermarkets), and schools or colleges.
When vaccines are given in nonmedical settings, it remains important for standards of immunization practice to be followed. Consumers should be provided with information on how to report adverse events (e.g., via provision of a VIS), and procedures should ensure that documentation of vaccine administration is forwarded to the primary care provider and the state or city public health immunization registry. Detailed documentation may be required for employment, school attendance, and travel. Personalized health records can help consumers keep track of their immunizations, and some occupational health clinics have incorporated automated immunization reports that help employees stay up-to-date with recommended vaccinations. Some pharmacy chain establishments are using automated systems to report immunization information to the state or local immunization information system.
PERFORMANCE MONITORING
Tracking of immunization coverage at national, state, institution, and practice levels can yield feedback to practitioners and programs and facilitate quality improvement. Healthcare Effectiveness Data and Information Set (HEDIS) measures related to adult immunization facilitate comparison of health plans. The CDC’s National Immunization Survey and National Health Interview Survey provide selected information on immunization coverage among adults and track progress toward achievement of Healthy People 2020 targets for immunization coverage. Influenza and pneumococcal vaccine coverage rates have been higher among persons ≥65 years of age (60–70%) than among high-risk 18- to 64-year-olds. Figures on state-specific immunization coverage with pneumococcal polysaccharide and influenza vaccines (as measured through the CDC’s Behavioral Risk Factor Surveillance System) reveal substantial geographic variation in coverage. There are persistent disparities in adult immunization coverage rates between whites and racial and ethnic minorities. In contrast, racial and economic disparities in immunization of young children have been dramatically reduced during the past 20 years. Much of this progress is attributed to the Vaccines for Children Program, which since 1994 has entitled uninsured children to receive free vaccines.
FUTURE TRENDS
Although most vaccines developed in the twentieth century targeted common acute infectious diseases of childhood, more recently developed vaccines prevent chronic conditions prevalent among adults. Hepatitis B vaccine prevents hepatitis B–related cirrhosis and hepatocellular carcinoma, zoster vaccine prevents shingles and postherpetic neuralgia, and HPV vaccine prevents some types of cervical cancer, genital warts, and anogenital cancers and may also prevent some oropharyngeal cancers (although this outcome was not studied in prelicensure randomized controlled trials). New targets of vaccine development and research may further broaden the definition of vaccine-preventable disease. Research is ongoing on vaccines to prevent insulin-dependent diabetes mellitus, nicotine addiction, and Alzheimer’s disease. Expanding strategies for vaccine development are incorporating molecular approaches such as DNA, vector, and peptide vaccines. New technologies, such as the use of transdermal and other needle-less routes of administration, are being applied to vaccine delivery.