Viruses
After reading this chapter, the student will be able to:
• Describe the two commonly used methods of classifying viruses
• Define and describe capsid, capsomeres, nucleocapsid, virion, and envelope
• Describe and differentiate the types of viral genomes
• Describe and explain the steps in the multiplication of bacteriophages and animal viruses
• Discuss the possible effects of viral infections
• Describe the different kinds of host cell damage caused by viral infections
• Name the DNA viruses infecting and subsequently causing diseases in humans
• Name the RNA viruses infecting and subsequently causing disease in humans
• Compare and contrast viroids and virusoids
• Describe prions and the infections/diseases caused by them
General Structure and Classification
Classification
For the determination of order, the type of nucleic acid DNA or RNA, single or double stranded, and the presence or absence of an envelope are used. In addition, other characteristics such as the type of host, the capsid shape, immunological properties, and the type of disease the virus causes are also considered for further detailed virus classification. An additional classification system is the Baltimore classification system, devised by David Baltimore. This system places the virus into one of seven groups distinguishing the viruses on the basis of the relationship between the viral genome and the messenger RNA (Box 7.1). In modern virus classification the ICTV system is used in conjunction with the Baltimore classification system. Although animal and plant virologists both use these systems, the actual application in the process of classification is quite different because of the diversity of viruses.
HEALTHCARE APPLICATION
Medically Important Viruses
| Family | Genus (or Subfamily) | Species or Typical Member | Infection/Disease |
| DNA Viruses | |||
| Parvoviridae | Erythrovirus | B19 virus | Erythema infectiosum |
| Papillomaviridae | Papillomavirus | Human papillomavirus (HPV) | More than 60 HPV types: common warts, plantar warts, flat cutaneous warts, etc. |
| Polyomaviridae | Polyomavirus | Polyomavirus | |
Morphology
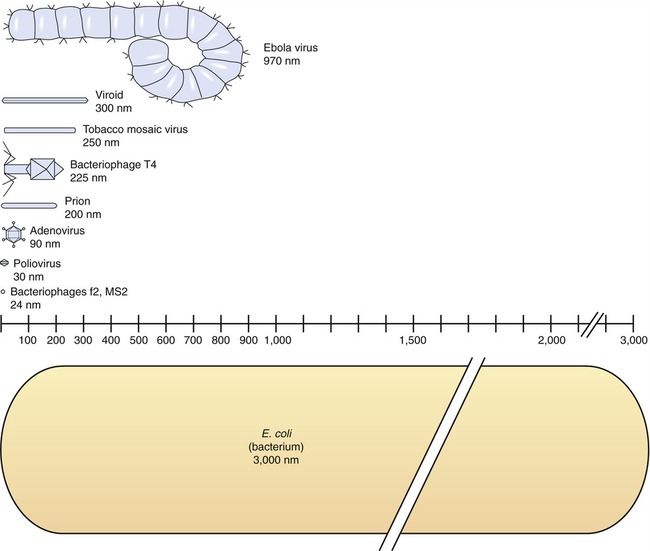
The size of viruses varies from very small, such as the parvovirus at about 20 nm and the poliovirus at 30 nm, to fairly large, such as the vaccinia virus at 400 nm and the poxviruses, which can be up to 450 nm. Most viruses cannot be visualized by light microscopy; therefore electron microscopy is used to examine their structure (see Chapter 1, Scope of Microbiology). However, some viruses are as large as or larger than the smallest bacteria and can be visualized by high optical magnification (Figure 7.1).
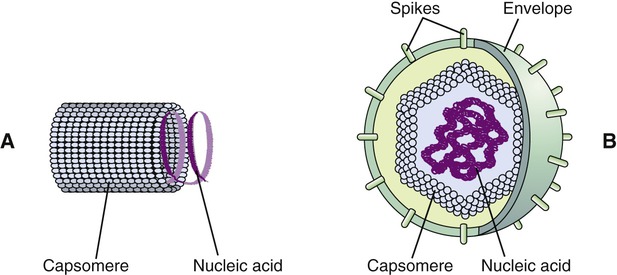
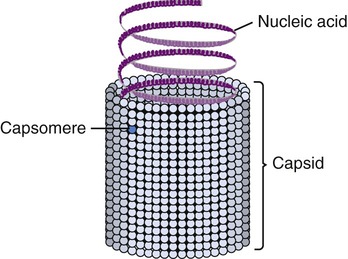
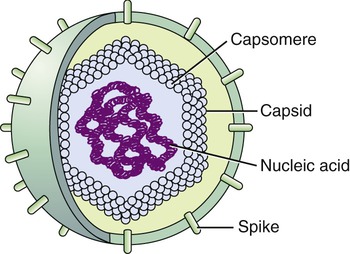
Viruses consist of genetic material carried in a “shell” called the viral coat or capsid. The capsid consists of proteins that are coded by the viral genome. The capsid is a complex and highly organized entity that gives form to the virus and serves as the basis for morphological distinction. Subunits referred to as protomeres assemble to form capsomeres, which in turn spontaneously aggregate to form the capsid. Proteins associated with nucleic acids are called nucleoproteins and the association of viral capsid proteins with viral nucleic acid is referred to as a nucleocapsid. At the junction points of the capsomeres, long projections from the nucleocapsids, called spikes, are frequently attached (Figure 7.2). These spikes aid in attachment to the host cell membrane receptors. Some viruses have an envelope around the coat and once the virus is fully assembled it is called a virion. Depending on the presence or absence of an envelope, viruses are referred to as naked or enveloped viruses (Figure 7.3).
Helical Viruses
Helical capsids have rod-shaped capsomeres, connected along their long axis, resembling a wide ribbon stacked around a central axis forming a helical structure with a central cavity, a hollow tube. As a result, these virions are rod shaped or filamentous and can be short and highly rigid, as in many plant viruses, or long and very flexible, as in many animal viruses. The genetic material can be single-stranded RNA (ssRNA) or single-stranded DNA (ssDNA) and is bound into the protein helix by the interactions between the negative charge on the nucleic acid and the positive charge on the proteins. The nucleocapsid of a naked helical virus is rigid and tightly wound into a cylinder-shaped structure. An example is the well-studied tobacco mosaic virus, an ssRNA virus that infects primarily tobacco plants and other members of the Solanaceae family (Figure 7.4). Enveloped helical nucleocapsids are more flexible and this type is found in several human viruses such as influenza and measles viruses.
Icosahedral Viruses
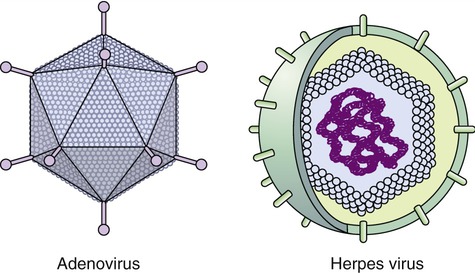
Capsids of many virus families are multifaced structures known as icosahedrons. These are three-dimensional, geometric figures with 12 corners, 20 triangular faces, and 30 edges (Figure 7.5). This icosahedral capsid symmetry results in a spherical appearance of viruses at low magnification. The arrangement of the capsomeres varies between the viruses and the shape and dimension of the icosahedron depend on the characteristics of its protomeres. Although the basic symmetry of icosahedral viruses is the same there are major variations in the number of capsomeres, and therefore differences in the size of the viruses. The basic triangular face of a small virus is constructed of 3 protomeres, with 60 of these subunits forming the whole capsid. For example, the poliovirus has 32 capsomeres, whereas an adenovirus has 240 capsomeres. During the assembling of an icosahedral virus the nucleic acid is packed into the center, forming a nucleocapsid. Icosahedral viruses can be naked, such as the adenovirus, or enveloped, as the herpes simplex virus (Figure 7.6). Included in this morphological group are the Iridoviridae (infect mainly invertebrates), Herpesviridae, Adenoviridae, Papovaviridae, and Parvoviridae.
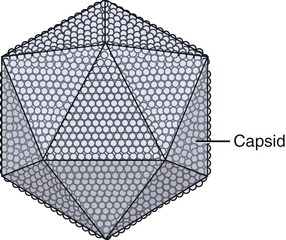
The capsids of these viruses are called icosahedrons: three-dimensional, geometric figures with 12 corners, 20 triangular faces, and 30 edges.
Enveloped Viruses
Many viruses have an outer structure, the viral envelope, which surrounds the nucleocapsid. Enveloped viruses typically obtain their envelope by budding though a host plasma membrane (see Plasma Membrane in Chapter 3, Cell Structure and Function) but may also include some viral glycoproteins. In some cases the viral envelope is derived from other membranes in the host cell, such as from the membranes of the endoplasmic reticulum or the nuclear membrane. The creation of the viral envelope through the budding process allows the viral particles to leave the host cell without disrupting the plasma membrane and therefore without killing the cell. As a result, some budding viruses can set up persistent infections.
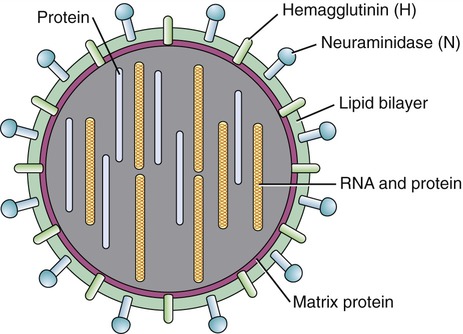
During the formation of the envelope some or all of the host membrane proteins are replaced by viral proteins and some form a layer between the capsid and the envelope. These envelope proteins are glycoproteins; they are exposed to the outside of the envelope as spikes. The lipid bilayer of the envelope is exclusively host specific whereas the majority or all of the glycoproteins are virus specific. The glycoproteins on the surface of the envelope serve to identify and bind to receptor sites on the plasma membrane of the host. Parts of the viral capsid and/or envelope are also responsible for stimulating the immune system of the host to produce antibodies that can neutralize the virus and prevent further infection (see Chapter 20, The Immune System). The viral envelope can give a virion protection from certain enzymes and chemicals; however, because enveloped viruses depend on their intact envelope to be infectious, agents that damage the envelope, such as alcohol and detergents, greatly reduce the infectivity of the virus (see Chapter 19, Physical and Chemical Methods of Control). An example of an enveloped virus is the influenza virus shown in Figure 7.7.
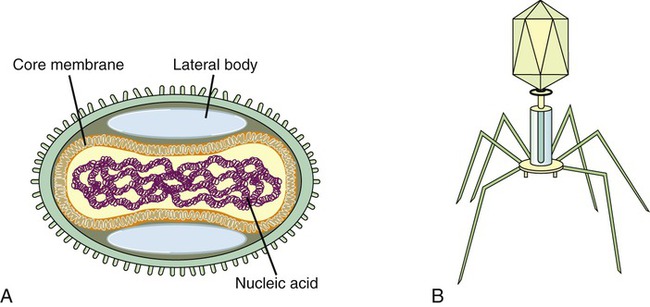
Complex Viruses
Complex viruses are a special group of viruses that consist of a capsid that is neither purely helical nor completely icosahedral and that has extra structures such as protein tails or a complex outer wall. The poxviruses are large, complex DNA viruses with an unusual morphology. These viruses lack a regular capsid and the viral genome is associated with proteins within a central disk structure, the nucleoid, which is surrounded by a membrane and two lateral bodies. The nucleoid is surrounded by several layers of lipoproteins and a coarse surface of fibrils (Figure 7.8, A).